This is the latest version of this chapter. The previous version can be found here.
Fakhar Abbas 1, Joseph C. Wu 2,3, Sanjiv Sam Gambhir 2,4, Martin Rodriguez-Porcel 1,§
1 Department of Cardiovascular Medicine, Mayo Clinic, Rochester, MN, USA
2 Molecular Imaging Program at Stanford, Stanford University, Stanford, CA, USA
3 Department of Medicine (Cardiology), Stanford University, Stanford, CA, USA
4 Department of Bio-Engineering, Stanford University, Stanford, CA, USA
Regenerative medicine with the use of stem cells has appeared as a potential therapeutic alternative for many disease states. Despite initial enthusiasm, there has been relatively slow transition to clinical trials. In large part, numerous questions remain regarding the viability, biology and efficacy of transplanted stem cells in the living subject. The critical issues highlighted the importance of developing tools to assess these questions. Advances in molecular biology and imaging have allowed the successful non-invasive monitoring of transplanted stem cells in the living subject. Over the years these methodologies have been updated to assess not only the viability but also the biology of transplanted stem cells. In this review, different imaging strategies to study the viability and biology of transplanted stem cells are presented. Use of these strategies will be critical as the different regenerative therapies are being tested for clinical use.
Over the last decade, regenerative medicine has appeared as a powerful alternative for the treatment of many diseases [1–4]. The main objective of cell-based therapies is to repopulate the damaged tissue with functional cells, with the final goal that these cells will integrate with the remaining functional native cells and contribute to the recuperation of the lost function. Regenerative medicine has been used to repair different organs/systems: endocrine (i.e., pancreas) [5], musculoskeleton (i.e., bone, joints) [6, 7], and cardiovascular system (i.e. myocardium) [8–10], as well as adjuvant treatment for malignancies [11]. While much has been learned on how stem cells function in cell culture, several questions remain regarding the biology of stem cells in living subjects. For most therapeutic applications of regenerative medicine, critical issues such as stem cell type and delivery route of stem cells remain to be elucidated. In addition, after cells are transplanted to the living subject, it becomes critical to understand the biology of transplanted cells and how they interact with the microenvironment. To answer some of these questions, it is imperative to perform these studies directly in the living subject in a longitudinal manner. Until recently those could not be performed in a reliable and accurate manner. Recent developments in molecular imaging modalities may likely permit investigators to answer some of these questions. Furthermore, some of these imaging strategies have the potential to be translated to patients, which makes them plausible to be used in the clinics.
REQUISITES FOR THE IDEAL IMAGING MODALITY FOR STEM CELL TRACKING
The ideal imaging agent/modality should provide the following information:
-
Real-time visualization of stem cell delivery
-
Determination of location(s) of cells over time
-
Quantification of numbers of viable transplanted stem cells
-
Long-term quantification of transplanted stem cell survival
-
Study of stem cell biology:
-
Interaction between stem cells
-
Interaction of stem cell with its microenvironment
-
Differentiation capacity of stem cells
-
The chosen labeling modality should not interact with the normal functions of the stem cell. Otherwise, one would not be able to accurately study the biology of these cells over time. In addition, issues such as biocompatibility, toxicity, and safety not only to the stem cell but most importantly to the individual should also be considered and included in the decision of which modality to use.
All imaging modalities have a certain degree of background/non-specific signal (that may interfere with the signal under study). The preferred imaging modality should be one that provides a good contrast between background and the target signal under study, achieving a large signal-to-noise ratio. Furthermore, it should have good specificity (negative study in the absence of what is being studied, stem cells in this case). Only then will it be possible to use these modalities to study stem cell biology.
The main objective of regenerative medicine, and thus of stem cell imaging, is its clinical applications. It is generally agreed upon that before its clinical use, therapeutic strategies should be tested in clinical models of disease. Thus, an ideal imaging modality should be flexible across different imaging modalities, both in terms of spatial resolution and system sensitivity (the lowest amount of activity or numbers of cells that can be detected by that specific modality).
METHODOLOGIES FOR LABELING STEM CELLS
Direct labeling
One of the most commonly used strategies for the labeling of stem cells for imaging in living subjects is that of direct labeling [12–15]. In a direct labeling strategy, labeling agents are introduced into the cells prior to transplantation, stem cells transplanted and then followed in the living subject (Fig. 1). The strategy is then used to image the molecules previously introduced into the cell and use them as surrogate of the number of stem cells. Depending on the imaging modality to be used, cells can be labeled with quantum dots (Fig. 1A) [16–18] or fluorophores [19–21] for optical fluorescence imaging, radionuclides for single photon emission computed tomography (SPECT) or positron emission tomography (PET) [22–25] and superparamagnetic iron oxide particles (SPIO, Fig. 1D) for magnetic resonance imaging (MRI) [26, 27].
Fig.1 Direct cell labeling. In a direct labeling strategy, labeling agents are introduced ex-vivo, stem cells are then transplanted to the tissue/organ of interest, and then non-invasive imaging is performed. Direct labeling can be achieved with fluorescence (quantum dots, QD, panel A), single photon emission computed tomography (SPECT) imaging 111-Indium (111In, panel B), positron emission tomography (PET) for the imaging of 18Fluorine-Fluorordeoxyglucose (18F-FDG, panel C) or magnetic resonance imaging (MRI) using superparamagnetic iron oxide particles (SPIO, panel D). Reprinted from Chen IY, et al. Cardiovascular Molecular Imaging: Focus on Clinical Translation. Circulation 2011; 123(4): 425-443 with permission.
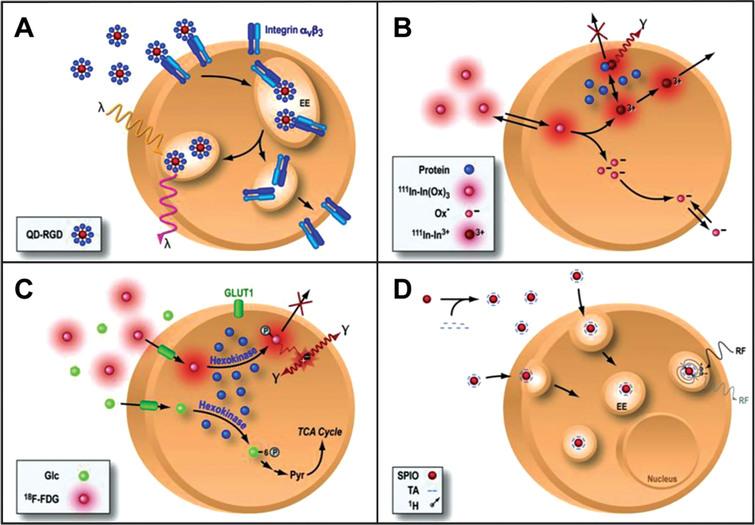
Fluorescent semiconductor nanocrystals (known as quantum dots, QDs [28] or different fluorophores [29] have been used to monitor stem cells in the living subject. At each imaging time point, QDs and fluorophores are excited at predetermined wavelengths, and emit a fluorescent signal that can be used for imaging of transplanted cells. The QDs have narrow emission spectrum and broad excitation spectrum and have demonstrated high resolution, long duration, high sensitivity [30]. This strategy has been used for tracking of stem cells in many organs [18, 28, 29, 31, 32]. However, fluorescence imaging has limited tissue penetration (around 2 mm) limiting the use of these techniques to superficial tissues in small animals (e.g., mice) [33, 34]. Several efforts are underway to provide tomographic fluorescence imaging [35–37]. More recently, there has been significant interest in the use of time domain imaging (TDI) as a way to identify the location of the detected signal [38]. In TDI, a specific location on the subject is illuminated with a laser pulse and a detector measures the arrival distribution of photons as a function of time at different locations. If one knows the amount of time it takes for the pulse to occur (typically sub nanosecond) and the time of arrival of the signal to the detector, one can infer the depth of the signal origination. Furthermore, TDI systems can provide temporal information from which fluorescence lifetime can be derived, allowing simultaneous imaging of fluorophores with different lifetimes even if they have overlapping spectra [39].
In this review, focus will be placed mainly on direct labeling modalities that have the potential to be applied clinically: MRI, SPECT, and PET.
Magnetic resonance imaging
Magnetic resonance imaging has been commonly used for imaging of stem cells, mostly based on concept of the imaging of SPIOs. SPIOs are highly magnetic particles that can elicit changes in T2 relaxivity (effect known as T2*) [40–45], allowing their detection in vivo (Fig. 1D). In a direct labeling strategy, SPIOs are incorporated into cells (using either electroporation or liposome-based incorporation techniques), cells then transplanted to the living subject, and subsequently MR imaging is performed [33, 46] using gradient recalled echo sequences [47]. The signal originated from the SPIOs is used as a surrogate for number of cells. MRI offers the advantage of high spatial resolution, resulting in detailed organ morphologic and functional information, and thus appears as a good candidate for an integrated stem cell imaging-functional assessment imaging approach in organs like the heart [46, 48] or the brain [14, 49]. MR has also been used to monitor the delivery process. Specifically, MR fluoroscopy allows real-time assessment of the delivery of stem cells to the myocardium [46, 48]. However, the sensitivity of SPIO-based labeling is in the micromolar range (10–5 mol/L) [33, 34] and may not be sensitive enough to detect low signal levels (more than 1×105 cells are needed) [50]. Sensitivity of the system can be increased using high-field magnets (e.g., 11 Tesla). However, for now these magnets are limited to use in small animal preclinical models. In recent years Ferrumoxytol, an ultra-small superparamagnetic iron oxide nanoparticle (USPION), has recently been used to track the fate of transplanted stem cells using magnetic resonance imaging (MRI) and has shown promising results. USPION are magnetic nanoparticles that have an active surface coating for different applications. Because Ferumoxitol is already approved by the FDA, albeit for a different indication, it has the potential to be used in future cardiac stem cell therapy trials [51–54].
Iron oxide-based imaging constitutes a very good imaging strategy for initial localization of cells after transplantation, and for the co-registration of cell transplantation with areas of functional loss. After myocardial infarction, MR imaging is used to identify areas of infarction (using delayed enhancement MR techniques), and SPIO-MRI can be used to label stem cells for their localization immediately after delivery [46] (Fig. 2 A). However, SPIO-based imaging is not well suited for long-term monitoring of stem cells [33, 47, 55]. Several reports suggested that SPIOs may not stay in the transplanted cells over time [55], but rather be incorporated by macrophages and other cell types, and some iron may remain in the interstitial space.
Fig.2 Imaging after direct cell labeling. Panel A, cells have been loaded with paramagnetic particles (Ferumoxitol), delivered to the brain, and then imaged using MRI. The black signal (red arrow) represents the paramagnetic signal. Panel B, rat cells were loaded with 111Indium Oxine ex-vivo, transplanted to the myocardium of rats, and then imaged using SPECT. SPECT has the capability of doing dual isotope imaging (e.g. 111InOx to image cells-red arrows- and 201Tl to image tissue perfusion-yellow arrows-). In panel C, bone marrow cells have been loaded with 18F-Fluorodeoxyglucose, delivered intracoronary to the myocardium, and then imaged using PET. The white arrows point to the transplanted cells in the heart, as well as the liver and spleen uptake (route of elimination of 18F-FDG). LV: left ventricle. Reprinted from Zhou R, et al. In Vivo Detection of Stem Cells Grafted in Infarcted Rat Myocardium. J Nucl Med 2005; 46:816-822, Hofmann M, et al. Monitoring of Bone Marrow Cell Homing into the Infarcted Human Myocardium. Circulation 2005; 111(17):2198-202, and Lee NK, et al. Magnetic Resonance Imaging of Ferumoxytol-Labeled Human Mesenchymal Stem Cells in the Mouse Brain. Stem Cell Rev and Rep 2017; 13:127-138 with permission.
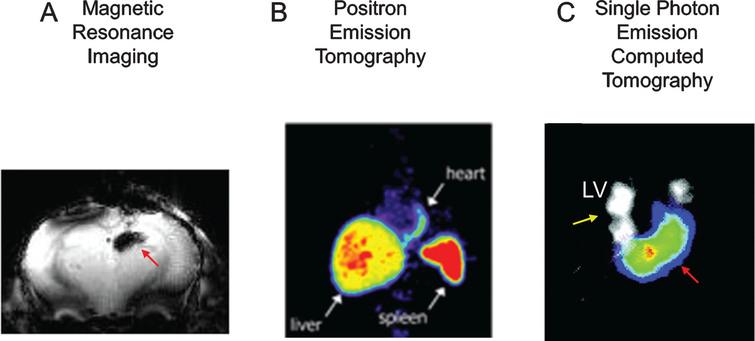
Because the effect of iron particles on the magnetic field continues (regardless of the location and status of the transplanted cells), there may be an uncoupling between the MR signal and the viability of stem cells [55]. This effect may not be critical for the initial localization of transplanted cells; but will preclude the use of this strategy for the monitoring of transplanted stem cells over time.
Radionuclide imaging
Radionuclide labeling of cells has also been used for cell imaging, using a strategy similar to SPIO-based techniques, which is to introduce a labeling agent to the cell prior to transplantation (Fig. 1B and 1C). Radionuclides used for this purpose can have different physical half-lives (e.g., 99mTc: 6 hours, 111In: 2.8 days, 18F: 109 minutes, 64Cu: 12 hours), that will determine the amount of time that cells can be monitored non-invasively after cell labeling. For example, 111In-labeled cells have been used for many years to track the homing of inflammatory cells to localize inflammatory processes [13, 56]. More recently, the methodology has been applied to the labeling of stem cells, using different isotopes (e.g., 111In for SPECT, 18F-Fluoro-Deoxyglucose (18F-FDG) for PET). Use of isotopes like 18F-FDG (physical half-life = 109 minutes) may allow tracking of cells for a 6–8 hours (after correction for isotope physical decay) after transplantation [22] (Fig. 2B) [57], while use of 111In (Fig. 2C) may allow cell tracking longer periods of time (up to 14 days) [58]. In addition to its physical half-life, each radionuclide also has a biological half-life (e.g., radionuclides may go in and out of the cell), which should be taken into consideration when performing these studies. More recently, there has been significant interest in the use of probes that have longer half-life (Zerconium, 89Zr) [59, 60]. One of the major advantages of SPECT and PET imaging is their high sensitivity (nano- and femto-molar, respectively), which permits the detection of relatively low amounts of signal [24, 61, 62]. However, SPECT and PET have relatively low spatial resolution, compared to other modalities (such as MRI), which may be a relative disadvantage for signal localization. The recent development of integrated PET-Computed Tomography (CT) and SPECT-CT provides a better anatomical guide for the location of the detected signal.
Advantages and disadvantages of direct labeling. Role in the assessment of stem cell therapy
Use of SPIOs (at least at doses of 20 pg of SPIO/cell) may have an effect on the gross morphology and proliferation capacity of stem cells, which may preclude their widespread use as a labeling strategy [43, 47]. In the case of radionuclide labeling, cell toxicity will likely vary depending on the radionuclide and dosage used [63, 64].
In addition, both SPIOs and radionuclides have a biological half-life (e.g., may go in and out of the cell), which should be taken into consideration when performing these studies. Thus, when using this imaging strategy, physical as well biological properties of the labeling agent should be considered together with the properties of the cells, to accurately determine the appropriate cell imaging modality. In general, the shortest half-life (biological or physical) will determine for how long transplanted cells can be monitored.
One of the main limitations of any direct labeling strategy is that it does not account for cell viability/division (i.e., cell numbers increase after cell division, but the number of radioisotope molecules stays the same), which results in “dilution” of the signal over time. This aspect of direct labeling limits its use for long term monitoring.
In summary, direct labeling strategies (whether it is MRI, SPECT-CT or PET-CT) appear to be a good imaging strategy for detection of cells shortly after transplantation (e.g., to ensure that cells where delivered to the intended organ or region of an organ), providing a good signal-to-noise ratio, but less suited for long-term monitoring of stem cell viability.
Reporter gene imaging
There is increasing need to understand the biology of stem cells after they are transplanted to the living subject. To achieve that, one needs a strategy that can accurately monitor stem cell biology in a longitudinal manner, without interfering with the normal biology of the cells under study or of the transplant recipient. In order to accurately study the biology of stem cells, such a system should be based on the physiologic activity of transplanted cells. Advances in molecular biology and imaging modalities have resulted in the development of reporter gene strategies, a system that allows evaluation of trans-gene expression in many disease states (Fig. 3) [34, 61, 65–71]. Reporter genes consist of gene regulatory elements (promoters and enhancers) that drive the reporter gene DNA sequence, and a polyA sequence (which provides stabilization to the final product). Initially, reporter genes were mainly used in histology and ex-vivo studies (beta-galactosidase and luciferase) to assess the trans-gene expression in many different tissues and organs [72–74]. Subsequently, reporter genes were used for imaging in vivo, using green fluorescent protein (GFP) [75] or bioluminescence (Fig. 3A) [65], providing one of the first evidences of the monitoring of trans-gene expression assessed in living subjects [76]. For the application of stem cell monitoring, the reporter gene is incorporated into the cell before cell transplantation into the living subject. If the stem cells are viable (e.g., after transplantation), the reporter gene will be expressed and the protein (e.g., enzyme, cell surface receptor) will be encoded. On the other hand, if the reporter gene is not expressed due to cell death for example, no signal will be produced. At the specified imaging time point, an exogenously given substrate is administered. The interaction between the substrate and the encoded reporter protein (if present) will result in a signal, which can be detected non-invasively (using different imaging modalities, e.g. bioluminescence imaging, radionuclide imaging). The most common use of reporter genes is for the longitudinal study of stem cell viability. For this purpose, reporter genes are driven by a constitutive promoter (e.g., cytomegalovirus or CMV), which is always turned ON and, as long as the cell is viable and has the transcriptional machinery intact, will result in production of the reporter protein. However, the viral CMV promoter can undergo “gene silencing” (the gene is turned OFF) over time [77] (a proposed mechanisms is linked to the large number of CpG repeats in the CMV promoter) that will result in reduced signal. Practically, a decrease in the activity of the constitutive CMV promoter will result in decreased production of the reporter gene which could be mistaken for decreased stem cell survival. More recently, investigators have used of promoters of constitutive mammalian cell proteins (e.g., β-actin, ubiquitin), which are proteins that are constantly produced in a cell. Most of these promoters undergo less gene silencing and stay “ON” as long as the cell is viable and may represent better alternatives as the promoter of choice for assessment of stem cell viability. Furthermore, cells should be monitored for alterations in their phenotype and dividing capacity, although recent data suggest that the introduction of reporter genes do not seem to significantly alter the biological properties and differentiation capacity of stem cells [78–80].
Fig.3 Different types of reporter gene/reporter probe strategies. (A) Enzyme-based bioluminescence imaging. D-Luciferin is a substrate molecular probe that is acted upon by the enzyme Firefly Luciferase to result in bioluminescence via a chemiluminescent reaction under physiological conditions only within living cells expressing the firefly luciferase (fluc) gene. (B) Enzyme-based PET imaging. transgene expression of a mutant herpes simplex virus type 1 thymidine kinase (HSV-sr39tk) reporter gene leads to the thymidine kinase enzyme (HSV1-sr39TK), which phosphorylates the PET reporter probe 9-(4-18F-fluoro-3-hydroxymethylbutyl)guanine (FHBG) and traps it intracellularly. Radioactive decay of 18F leads to positron (+) emission and subsequent annihilation with a nearby electron (–) to produce 2 oppositely directed gamma rays as cell signal C) Receptor-based SPECT imaging. expression of sodium iodide symporter (NIS) reporter gene leads to insertion of sodium iodide symporters into the cell membrane, where they import either 123I– or technetium pertechnetate (99mTcO4–) as reporter probe, along with sodium ion (Na+), into the cytosol. Imagig is performed with SPECT D) MRI: transgene expression of either ferritin heavy chain (FTH) or ferritin light chain (FTL) MR reporter gene leads to the assembly of ferritin (FT) proteins, which can sequester intracellular iron (Fe). The paramagnetic properties of iron allow them to be imaged with MRI. Reprinted from Cardiovascular Molecular Imaging: Focus on Clinical Translation. Chen IY, Wu JC. Circulation 2011:123(4): 425-443 with permission.
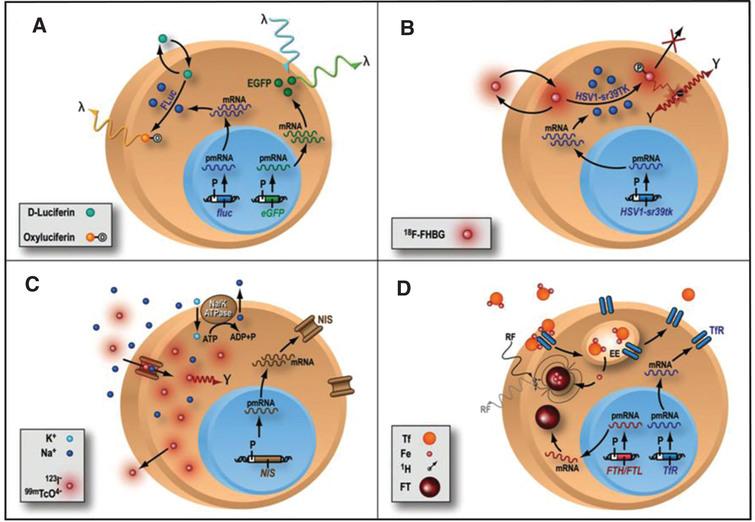
Specific biological pathways can also be investigated using reporter genes strategies. To study a certain pathway, a specific promoter is used (e.g., a protein specific promoter), and only when the intracellular signal for the production of that protein is active, would the reporter protein be made. For example, if one wants to monitor when embryonic stem cells differentiate into myocytes, one can use a reporter gene that is driven by a specific myocyte promoter, which will only be turned “ON” when the stem cell under study has turned “ON” the signaling cascade to produce a mature cardiac protein (e.g. troponin, desmin). Then, the activation of that specific promoter will “drive” the expression of the reporter gene, and one could visualize it non-invasively using different imaging modalities.
Fluorescent reporter genes
Use of GFP constitutes one of the first examples of the application of reporter genes for in vivo imaging of trans-gene expression. Fluorescence reporter proteins are very sensitive and result in the emission of a strong signal. However, both light excitation and emission undergo significant tissue attenuation (due to absorption) and tissue refraction, which limits the tissues that can be studied (up to 2 mm in depth) [34, 81, 82]. In view of this, the most used application of fluorescent reporter genes in stem cell imaging is for ex-vivo analysis, where they can be used for cell sorting. An issue to be considered in fluorescence imaging is that it is mainly a planar imaging technique, with no tomographic capabilities. Novel developments, such as time-domain imaging [83–85], are incorporating the time domain in the analysis, and have the potential to provide depth information of the fluorescent signal. In addition, new amplification system like protein scaffold e.g. sung-tag have been developed which can recruit multiple copies of GFP or transcription enhancing factors leading to amplified production of florescent reporter protein like GFP and considerably enhance the signal to overcome weakening of signals caused by increased tissue depth and the results are promising [86]. Another common application of fluorescent reporter genes is for use in histology (immunofluorescence) for post-mortem confirmation of imaging results [87]. One can “mark” cells with a fluorescent reporter gene, and then when the tissue is excised, the “marked” cells can be easily identified using histological methods.
In a recent study a new application developed with florescent reporter genes in induced pluripotent stem cells derived cardiomyocyte (hiPSC-CM) was the expression of genetically encoded voltage (ArcLight) and calcium (GCaMP5G) fluorescent indicators. Expression of ArcLight and GCaMP5G in hiPSC-CMs permitted to reliably follow changes in transmembrane potential and intracellular calcium levels, respectively [88].
Bioluminescent reporter genes
Reporter gene-bioluminescence imaging (BLI) is based on light emission and detection by specific cooled charge coupled device (CCD) cameras (Fig. 4A) [65]. Similar to other reporter gene strategies, the BLI signal is only emitted when cells are viable, and thus can be used for the longitudinal monitoring of stem cell survival and study of cell status (Fig. 4A). BLI is commonly used for the tracking of stem cells delivered to organs within small living animals [89–91]. Fluc and Renilla luciferase (Rluc) are the two most common reporter genes used for BLI. BLI-Fluc imaging (Fig. 4A) is based on the oxidation of the substrate D-luciferin by the FLuc enzyme, a reaction that requires oxygen, magnesium, and ATP, and results in a red-shifted light emission (wavelength: 500–700 nm). Imaging in the red-shifted light spectrum results in higher signal-to-background ratio, with less attenuation by red blood cells as well as improved tissue penetration with increases the amount of signal that leaves the body and increasing signal detection. Because of all these factors, it is more “attractive” as a reporter gene alternative for imaging in living subjects. On the other hand, Rluc does not require other cofactors and result in a lower wavelength emission (wavelength: 450–550 nm), resulting in a lower signal-to-background ratio that makes it more challenging for imaging in the living subject. More recently, the Gambhir lab has developed red-shifted Rluc variants that have increased light output which can make Rluc a more attractive option for in vivo animal imaging [92, 93].
Fig.4 Examples of reporter gene imaging. Panel A, shows the different imaging modalities used in reporter gene imaging, such as optical bioluminescence imaging (left), and radionuclide medicine imaging (PET-right). Subjects are placed in the cooled CCD camera system or scanner, respectively, and the substrate administered. Panel B presents examples of cell imaging using a reporter gene approach using the previously mentioned modalities. On the left panel, cells are genetically modified to carry the optical fluc reporter gene, delivered to the myocardium, and also imaged using a CCD camera after administration of the substrate D-Luciferin. Color images of visible light are superimposed on photographic images of mice with a scale in photons per second per square centimeter per steradian (sr). On the right panel, cells carrying the PET reporter gene HSV1-tk have been transplanted to the myocardium. The figure depicts tomographic images of the myocardium after the administration of the PET reporter probe 18Fluorine 9-[4-fluoro-3-(hy{droxy methyl)butyl]guanine (18F-FHBG). The color scale (% ID/g) indicates the percentage of injected dose that accumulates per gram of tissue. Red arrows indicate the area where cells are located. Reprinted from Cao F, et al. In Vivo Visualization of Embryonic Stem Cell Survival, Proliferation, and Migration After Cardiac Delivery. Circulation 2006; 113(7):1005-1014 and Lijkwan MA, et al. Trends Cardiovascular Medicine 2010; 6:183-8 with permission.
![Examples of reporter gene imaging. Panel A, shows the different imaging modalities used in reporter gene imaging, such as optical bioluminescence imaging (left), and radionuclide medicine imaging (PET-right). Subjects are placed in the cooled CCD camera system or scanner, respectively, and the substrate administered. Panel B presents examples of cell imaging using a reporter gene approach using the previously mentioned modalities. On the left panel, cells are genetically modified to carry the optical fluc reporter gene, delivered to the myocardium, and also imaged using a CCD camera after administration of the substrate D-Luciferin. Color images of visible light are superimposed on photographic images of mice with a scale in photons per second per square centimeter per steradian (sr). On the right panel, cells carrying the PET reporter gene HSV1-tk have been transplanted to the myocardium. The figure depicts tomographic images of the myocardium after the administration of the PET reporter probe 18Fluorine 9-[4-fluoro-3-(hy{droxy methyl)butyl]guanine (18F-FHBG). The color scale (% ID/g) indicates the percentage of injected dose that accumulates per gram of tissue. Red arrows indicate the area where cells are located. Reprinted from Cao F, et al. In Vivo Visualization of Embryonic Stem Cell Survival, Proliferation, and Migration After Cardiac Delivery. Circulation 2006; 113(7):1005-1014 and Lijkwan MA, et al. Trends Cardiovascular Medicine 2010; 6:183-8 with permission.](https://content.iospress.com/media/stj/2019/1-1/stj-1-1-stj190003/stj-1-stj190003-g004.jpg?width=755)
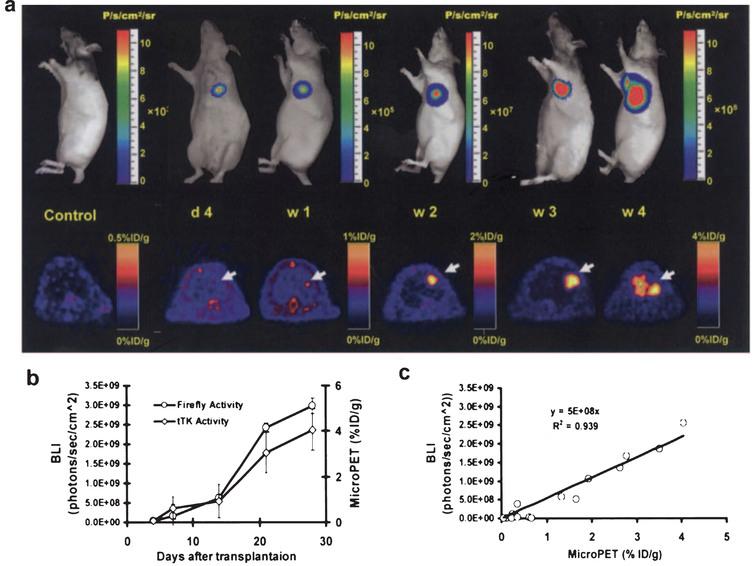
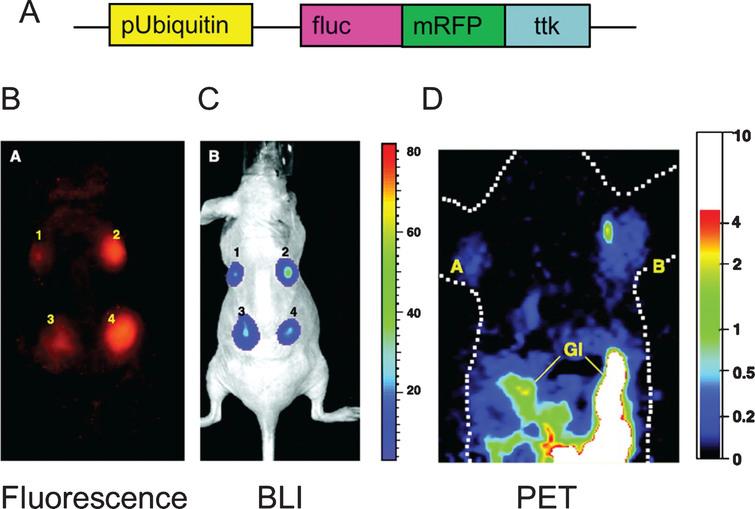
Using a reporter gene strategy, BLI has been successfully used for in vivo study of cell delivery and monitoring of stem cell viability in small living animals (Fig. 5). BLI has been used for tracking and monitoring of different types of cells, such as neural cells [94], cardiomyoblasts [47, 90, 91] and embryonic stem cells [95, 96]. For the delivery of stem cells to the myocardium, investigators have used ultrasound guidance. This strategy (ultrasound guidance for the delivery of stem cells) allows the precise delivery of stem cells to the affected part of the organ (myocardium in this case) [91, 97]. Furthermore, in these studies ultrasound guided delivery is complemented with non-invasive assessment of cardiac function (as it routinely done in the clinic). BLI has also been used to evaluate the response of stem cells transplanted to organs like the pancreas [98]. Furthermore, using reporter genes, Kutscha et al. have shown that modulation of the cellular and local microenvironment resulted in prolonged stem cell survival after transplantation to the myocardium [99, 100]. Our group has also shown that BLI has been used to show how the hostile ischemic myocardium negatively affects the biology of transplanted stem cells [101, 102].
Fig.5 Longitudinal monitoring of stem cell survival using reporter gene imaging. Molecular imaging of transplanted stem cells with bioluminescence and PET imaging. a, To assess longitudinal cell survival, animals were imaged for 4 weeks. A representative study animal injected with ES-TF cells showed significant bioluminescence (top) and PET (bottom) signals at day 4, week 1, week 2, week 3, and week 4. In contrast, control animals had background activities only. b, Quantification of imaging signals showed a drastic increase of fluc and ttk activities from week 2 to week 4. Extracardiac signals were observed during subsequent weeks. c, Quantification of cell signals showed a robust in vivo correlation between bioluminescence and PET imaging (r2 = 0.92). BLI indicates bioluminescence. Reprinted from Cao F, et al. In Vivo Visualization of Embryonic Stem Cell Survival, Proliferation, and Migration After Cardiac Delivery. Circulation 2006; 113(7):1005-1014 with permission.
As with other imaging modalities, BLI has some drawbacks and unsolved issues. Currently, BLI is mainly a planar imaging modality, providing limited depth information and signal localization within the living subject. Also, because of the lack of tomographic information, while BLI can provide information on the trafficking of stem cells to different organs, it may not be an ideal modality to accurately assess changes in stem cell viability as cells traffic between organs. This is because changes in the depth of the originated signal may be confused with changes in cell survival. Significant efforts are being devoted to the development of tomographic BLI either by rotating the subject under study, by detecting light using 2 or more cameras, or using spectral imaging [103, 104], To increase the signal to be detected, different groups have developed amplification systems [86, 105–108]. Recently conducted studies show that chemical modification of the luciferin substrate together with mutant luciferases extended the capabilities of bioluminescent reporters resulting in high photon flux in the red and near infrared spectrum which can help in studying the stem cells in deeper tissue and organs [109–111]. Due to the limited tissue depth that can be assessed with BLI, it is a technique that is mainly restricted to small animals (rats and mice), and very superficial tissues in larger living subjects (e.g., skin, in an intraoperative setting). In the future, these strategies may also be applied in specific clinical scenarios (e.g., skin, superficial tissues). Because BLI uses imaging strategies (reporter genes) similar to clinical modalities (e.g., PET and SPECT), BLI is routinely used as a starting step in the development of novel imaging strategies, and once the efficacy of these strategies is proven they can be adapted for use in the clinic.
In summary, BLI is a very useful imaging modality for the monitoring of stem cell trafficking and survival in small animals and will play a critical role in the study of cell biology and its interaction with the microenvironment.
Positron emission tomography/single photon emission computed tomography reporter genes
Both SPECT and PET reporter gene imaging are based on the interaction between an exogenously administered probe (that contains the tracer) and the reporter gene product (e.g., enzyme, receptor, Fig. 3B and 3C), what results in the retention of the radionuclide substrate that can then be imaged non-invasively [33, 61, 112–114]. One of the main advantages of PET and SPECT imaging is that it can provide tomographic, quantitative, and volumetric information, allowing one to better localize and quantify the detected signal within the subject under study (Fig. 4B). In addition, the sensitivity of PET is in the femtomolar range (10–12 mol/L), higher than MR imaging (10–5 mol/L), but not as sensitive as optical imaging (at limited depths, 10–15 mol/L). There are predominantly three reporter gene systems (for PET or SPECT) that have been used for cell imaging. The system used the most is based on the production of an intracellular enzyme (e.g., HSV1-tk), that phosphorylates an exogenously administered substrate which is retained in the cell due to its negative charge. While normal cells (without the HSV-tk) do carry the enzyme mammalian wild type thymidine kinase, it only minimally phosphorylates the radionuclide probes used in this system. On the other hand, in cells carrying the HSV-tk, the exogenously administered probe undergoes significant phosphorylation and intracellular retention, leading to a robust signal-to-background ratio, enabling accurate monitoring of these cells. Furthermore, this strategy is very powerful because the enzyme can phosphorylate many molecules of the radionuclide substrate, increasing the signal retained in the cells of interest and improving the signal-to-background ratio. However, the probe has to cross the cell membrane, which may limit the interaction between substrate and enzyme, resulting in reduced signal. In addition, because it is a non-mammalian protein (of viral origin) it has the potential to trigger an immunological response, resulting in a reaction in the organism with decreased overall signal. Attempts to circumvent this problem have included use of a mammalian protein (dopamine receptor, see later in this paragraph) or a mammalian mitochondrial tk [115], or a destabilized HSV1-tk [116]. A second reporter gene imaging strategy is based on the imaging of dopamine receptor using PET [117]. In this case, the reporter gene encodes for a cell membrane protein, which binds to an exogenously given probe, and the “bound probe” is then imaged non-invasively with PET. It is important to mention that the wild type D2R has the potential to elicit a downstream biological response, which can be prevented if one uses a mutant version of the dopamine receptor [89, 118]. Importantly, the mutant version of the D2R (Mutation of Asp80 or Ser194) maintains the affinity for the PET probe (3-(2’-[18F]-fluoroethyl)-spiperone) used. This strategy can be advantageous as the probe does not have to cross the cell membrane in order to interact with the reporter protein. However, this approach is limited by the amount of signal that can be produced, as one receptor interacts with only one molecule of the ligand. One of the most common strategies is the use of a PET-based reporter gene strategy for the visualization of transplanted cells to the myocardium and showed that it can be used to monitor the fate of stem cells after transplantation to the living subject [87, 90]. Similarly, we have shown that transplantation of pancreatic islet cells can be followed non-invasively [119–121], similar to what can be done with cancer imaging and treatment [122]. A third approach consists on the encoding of the sodium-iodine symporter (NIS, Fig. 4C) [123–126], which is a thyroid transmembrane protein, under physiological conditions transport iodine into the cells, in exchange for sodium. Successful sequencing and cloning of the NIS gene sequence allowed its use as a reporter gene in the thyroid and other organs. This system has been used not only for PET (with 124I as the tracer), but also for SPECT imaging (using 123I or 99Tc-perthechnetate as tracer). The somatostatin receptor reporter gene [127] can also be evaluated with both SPECT and PET as imaging modalities, providing flexibility on the assessment of trans-gene expression [128–130]. Other less frequent reporter genes include the neurotensin receptor subtypes [131, 132], and cytosine deaminase [133, 134]. Details of the use of these reporter genes are beyond the scope of this review and can be found elsewhere [128].
The major advantage of PET and SPECT in their high sensitivity and their ability to detect only live cells and possibility of using antivirals like ganciclovir in HSV1-tk model to be used as suicide gene to decrease the risk of tumorigenicity and dual use of NIS as an imaging and therapeutic modality [95, 122, 135]. Furthermore compared to fluorescence or bioluminescence, a major advantage of PET and SPECT resides in their perceived potential for clinical use. That being said, fluorescence-based agents have been used for the imaging of superficial tissues like skin and esophagus; underscoring its potential for clinical application [136, 137].
Both PET and SPECT radionuclides are of relatively high energy (PET: 511KeV, SPECT: 80–250 MeV) and do not undergo significant tissue attenuation. While SPECT (due to its relatively low energy) does have some tissue attenuation, it does not preclude its use in patients and proof of that is its extensive clinical use.
These imaging modalities present a number of issues that need to be considered from the operational standpoint. On one side, PET has significant flexibility for the production of specific probes for the detection of different processes in the living subject (almost any compound can be labeled with a radionuclide), which is a significant advantage as it allows the researcher to first identify the molecule that needs to be studied, and then design a specific probe that will target that molecule. However, the production of PET probes is complex, needs advanced chemistry and very tight quality control. In addition, depending on the half-life of the radioisotope used, it requires an on-site (or at least near-by) cyclotron, that limits this strategy to medium to large research centers. From the imaging standpoint, all electron-positron annihilations (whether is from 18F, 64Cu or 11C) result in the production of photons of 511KeV, and as such we cannot detect differences in registered signals. SPECT, on the other hand, can detect simultaneous signals of different energies (by varying the detection windows, as it is routinely done with the perfusion agents 201Tl and 99Tc). At the same time, tracer labeling is less complex (compared to PET) and, for the most part, can be done in a radionuclide pharmacy. However, the spatial resolution of SPECT is less than that of PET, and this variable may be of importance when we try to spatially localize relatively low number of cells. From the tracer perspective, SPECT labeling can be somewhat limited by its chemistry, with the result that not every compound can be easily labeled, thus providing less labeling flexibility compared to PET. In practical terms, if the compound of interest can be labeled with SPECT and answers the research questions posed by the investigator, its production and availability is more accessible to many academic and research centers. The other issue to be considered is the immunogenicity of these genes as they are derived from nonhuman protein and once they translated in genetically engineered therapeutic cells, there peptides are displayed on the cell surface, recognized by CD8+ cells, which attack and may kill the therapeutic cells [138].
In an effort to combine the benefits of the different imaging modalities (fluorescence, bioluminescence and positron emission tomography), the Gambhir’s laboratory and other groups have created a triple fusion reporter gene that has a fluorescent reporter gene, a bioluminescent reporter gene, and a PET reporter gene and that results in a fusion protein (Fig. 6) [139, 140]. For example, Cao et al. have transduced mouse embryonic stem cells, using the fluorescent protein (red fluorescent protein, mRFP) to identify the cells that have been effectively transduced with the fusion protein (using fluorescent activated cell sorting), and then transplanted those cells to the myocardium [95, 96]. Subsequently, the bioluminescent (firefly luciferase, Fluc) and PET reporter genes (herpes simplex virus enzyme thymidine kinase, HSV1-tk) were used for long-term monitoring of cell viability after transplantation [95, 96].
Fig.6 Triple fusion reporter gene. The design of the triple fusion reporter gene is depicted in panel A. The promoter Ubiquitin drives the expression of the fusion protein. fluc: Firefly luciferase (bioluminescence), mrfp: Red fluorescent protein (fluorescence), ttk: thymidine kinase (PET). The fluorescent protein (panel B) is use for fluorescent activated still sorting (FACS) ex vivo, while it can also be used for histology ex-vivo. In this study, stem cells carrying the triple fusion reported gene were delivered to the myocardium of rodents and imaged with the bioluminescence portion of the reporter gene with high sensitivity (panel C) while the positron emission tomography imaging capabilities of the reported gene is critical for clinical translation (panel D). Reprinted from Ray P, et al. Construction and Validation of Improved Triple Fusion Reporter Gene Vectors for Molecular Imaging of Living Subjects. Cancer Research 2007 Apr 1;67(7):3085-93 with permission.
Magnetic resonance reporter genes
Over the last few years, significant efforts have been devoted to combined these modalities (MRI and reporter gene technology) with the goal of developing MR reporter genes (Fig. 4D) [141, 142].
MRI-based reporter genes efforts are based on the production of proteins, mostly intracellular metalloproteins (transferrin, ferritin, tyrosinase) [142]. As previously described, iron is a paramagnetic substance that induces changes in relaxivity (i.e., T2* effect) that can be detected using specific imaging sequences. Physiologically, iron enters cells through the transferrin receptor (TfR) that binds the transferrin protein containing two iron atoms and internalizes iron molecules. So the goal of this strategy is to express transferring, that will accumulate large quantities of iron intracellularly for non-invasive detection.
A reporter gene strategy targeted to express Ferritin has also been used for MR-based detection. Ferritin is a metalloprotein that functions as the body’s iron depot and can contain up to 4000 iron atoms. Native ferritin is in essence an anti-magnetic particle, but several orders of magnitude weaker than SPIOs. Considerable efforts are being devoted to improve its relaxivity by removing its native core (oxyhydroxide) and reconstituting the protein shell with a superparamagnetic core. In another study Naumova et al. used mouse skeletal myoblast that were engineered to over express ferritin and transplanted them into infarcted hearts of the mice, not only the ferritin over expression did not interfere with cell biology and viability but also induced significant T2 relaxation that even after 3 weeks they were able to detect graft in the infarcted heart as an area of hypo intensity [143].
Tyrosinase has also been used as a MR reporter gene [144]. Briefly, tyrosinase participates in the production of melanin, and melanin has high affinity for iron, leading to increased relaxivity. Tyrosinase has been transfected to fibroblasts and embryomal kidney cells as well as breast cancer cells which resulted in increased signals. However, there is concern regarding potential toxicity of iron. Melanin production produces reactive oxygen species (an important and deleterious component of the oxidative stress cascade) and thus can exhibit significant toxic effects.
There are a few drawbacks in the use of metalloproteins as MR reporter genes that also deserve consideration [142]. First, the signal is dependent on the accumulation of iron inside the cells and for how long iron particles can be retained inside cells, what may result in toxicity to the cell. Second, when cells divide the signal gets diluted and the “clock” starts again, as cells need to start again to accumulate enough iron for MRI detection, making it difficult to interpret correlation between detected signal and viability of transplanted cells. From the imaging standpoint, the relaxation is dynamic and dependent on the iron loading conditions. R2 relaxivity is high at low iron doses, it decreases at intermediate iron loading conditions and when iron conditions are high, T2 relaxation remains constant, which may preclude accurate quantification of the obtained signal. As previously described for direct cell labeling, MR signal works on the basis of accumulation of iron. Similar to what happens with SPIOs, when cells die the accumulated iron continues to be present inside the cells for some time (until cells are dissolved or phagocytosed by macrophages). The remaining iron continues to induce a change in MR relaxivity (even if cells are not alive), and thus the signal is not representative nor linear of cell viability [55].
Translation to clinical applications
Over the last few years, the majority of our understanding of stem cells comes from studies in small animals. However, before these therapies are routinely applied clinically, there are a number of questions that need to be answered, such as dose, timing of delivery, homing, etc. For most of these answers to truly address the clinical dilemma, they need to be answered either in a large animal model or directly in the patient. The ability to directly monitor and assess cell-based therapies in patients will be invaluable as it will allow us for the first time to investigate these therapies directly in the living subject to which they were intended to. In many pathophysiological states, large animal models have been shown to be similar to human in what respects to disease progression [145]. These similarities have led researchers to use large animal models for diagnosis of disease as well as evaluation of different therapies.
In addition, many large animals have comparable weight, size, and anatomy to humans. These similarities also allow a better optimization of the different imaging strategies prior to clinical applications. In diseases where the use of large animal models is not relevant or feasible, the translation from small animal models to humans may be more difficult.
Recently, a number of research groups have used the swine model for imaging of reporter genes in the myocardium [146–148], showing the feasibility of applying these imaging strategies for gene monitoring in large animals. A similar strategy has been used for the imaging of stem cells (previously labeled to carry the reporter gene) in a large animal model (Fig. 7) [149] and humans (Fig. 8) [150]. However, it is important to realize that when cells are exogenously delivered to a host, a myriad of mechanisms may play a role in the survival of the transplanted stem cells: a relatively hypoxic scenario, lack of constant and efficient cell-cell contact with the microenvironment and activated immune response. In addition to the biological variables already mentioned, imaging of large animals (similar to clinical imaging) has a few technical aspects that need to be kept in mind. On one side, the sensitivity of clinical systems is lower than that of dedicated small animal imaging systems, which make imaging and signal quantitation more challenging. In addition and depending on the imaging modality used, the amount of the reporter gene/cell system to be delivered may need to be adjusted depending on the weight and other characteristic of the subject under study.
Fig.7 PET-CT imaging of intramyocardial reporter gene expression in swine. (a) Transverse nonenhanced PET-CT fusion image reconstructed at the level of the left ventricle (LV) after direct open-chest administration of transduced human MSCs. The image was acquired 4 hours after intravenous FHBG administration. A distinct imaging signal (small arrows) can be delineated at the intramyocardial injection site of human MSCs. Note postoperative soft-tissue edema, emphysema, and fluid collection in the left chest wall (large arrows). T = beveled part of chest tube. Reprinted from Willmann J, et al. Imaging Gene Expression in Human Mesenchymal Stem Cells: From Small to Large Animals. Radiology 2009 Jul;252(1):117-27 with permission.
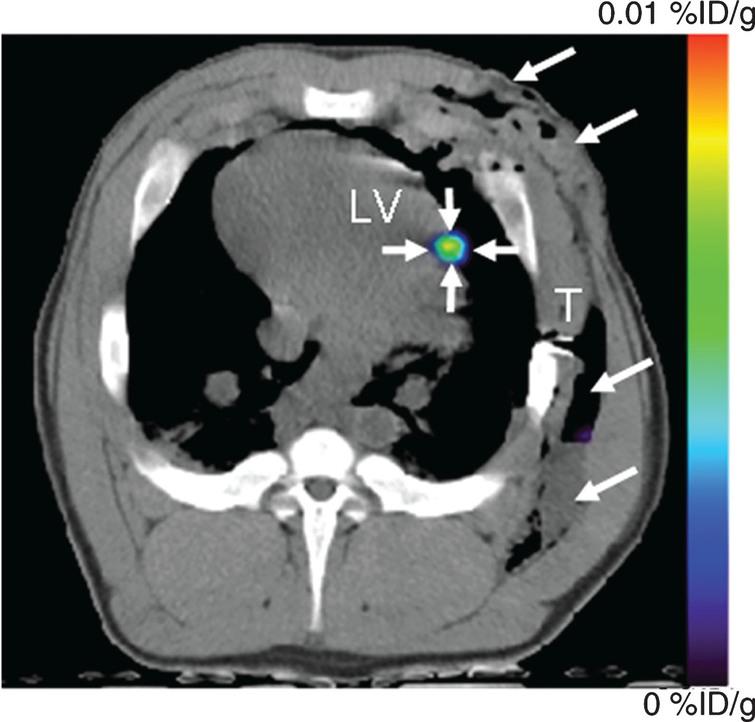
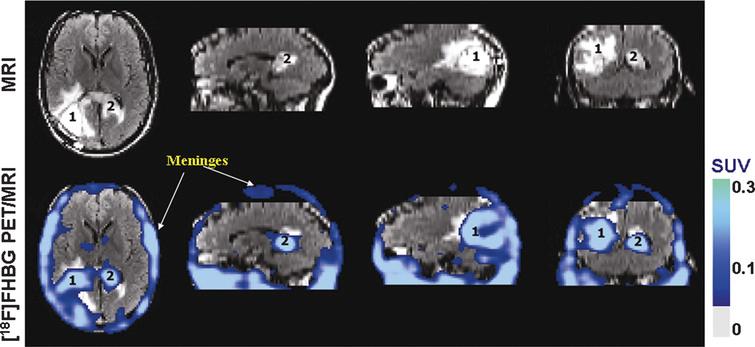
Fig.8 Human stem cell imaging. MRI and PET over MRI superimposed brain images of the patient who had been infused autologous cytolytic T cells expressing IL13 zetakine and HSV1-tk genes. Images were acquired approximately two hours after 18F-FHBG injection. The patient had a surgically resected tumor (1) in the left corner and a new non-resected tumor in the center (2), near corpus callosum of his brain. The infused cells had localized at the site of tumor 1 and also trafficked to tumor 2. 18F-FHBG activity is higher than the brain background at both sites. Background 18F-FHBG activity is low within the Central Nervous System due to its inability to cross the blood brain barrier. Background activity is relatively higher in all other tissues. Activity can also be observed in the meninges. The tumor 1/meninges and tumor 2/meninges 18F-FHBG activity ratio in this patient was 1.75 and 1.57, respectively. Whereas the average resected tumor site/meninges and intact tumor site to meninges 18F-FHBG activity ratio in control patients was 0.86 and 0.44, respectively. Reprinted from Yaghoubi S, et al. Noninvasive Detection of Therapeutic Cytolytic T Cells with 18F-FHBG PET in a Patient with Glioma. Nat Clin Pract Oncol 2009 Jan;6(1):53-8 with permission.
Clinical application
Direct labeling has already been achieved and tested in a clinical population [151–153] with varying success, not due to the safety of the methodology per se but rather due to the limitations of the direct labeling approach mentioned above. In other words, it has been very successful when used to identify where cells have been delivered, but not for long-term tracking. As examples, Kang et al. used 18F-FDG to label stem cells prior to transplantation [22], while other groups have used 111In [154, 155]. An example of MRI based imaging is the use of iron oxide for tracking of stem cells [153].
The application of reporter gene imaging to humans has several issues to be considered from the regulatory perspective. Initially, reporter gene approaches include the stable expression on the DNA cell using random integration, potentially leading to non-desired effects. However, development in genome editing with site-specific integration of DNA have made significant improvements in that respect [156–158], minimizing if not eliminating the above-mentioned concern. From the perspective of the reporter probes that are needed, different groups have been able to produce it in the FDA approved manner. In short, while it is feasible it requires very close collaboration among research groups and regulatory agencies.
How to decide which modality to use
The ideal imaging modality is one that has excellent spatial resolution and molecular sensitivity, can guide the delivery of cells, and can serially follow stem cells and their fate. Currently, no such imaging modality exists. Each imaging modality should be chosen depending on the question that is being asked (Fig. 9, Table 1). If the objective of the study is to image the delivery and short-term homing of stem cells in different organs, a direct labeling approach may answer the posed question, taking into consideration the potential toxicity that they may have. Magnetic resonance imaging provides the highest spatial resolution and almost real-time image-guidance for cell delivery, albeit with significantly lower molecular sensitivity compared to other modalities like PET, SPECT or optical imaging. Radionuclide imaging modalities (PET, SPECT) have been successfully and extensively used, although depending on the application they may not provide sufficient spatial resolution. Another research objective may be the long-term monitoring of stem cells viability. The drawbacks of a direct labeling strategy for stem cell monitoring have been previously described. On the other hand, reporter gene imaging appears more suited for the long-term monitoring of stem cells, and can be achieved using optical imaging (bioluminescence, fluorescence) or PET/SPECT imaging. While optical imaging is more molecularly sensitive, it provides lesser anatomical localization, and is limited mainly to small animals. On the other hand, PET or SPECT provide good sensitivity with the major advantage of good anatomical localization and potential translation to human applications.
Fig.9 Summary of the different imaging strategies that can be used to assess the delivery, short- and long-term monitoring of stem cell viability and biology. MRI: magnetic resonance imaging, SPECT: single photon emission computed tomography, PET: positron emission tomography, RFP: red fluorescent protein, GFP: green fluorescence protein, Fluc: firefly luciferase, Rluc: renilla luciferase, NIS: sodium iodine symporter, HSV1-tk: herpes simplex virus type 1 thymidine kinase, D2R: dopamine receptor type 2, CD: cytosine deaminase.
Table 1 Comparison of the spatial resolution and molecular sensitivity of the different imaging modalities discussed
| Monitoring Strategy | Spatial Resolution | Cell Detection Sensitivity |
| Direct Labeling | ||
| Fluorescence | 2+ | 3+ |
| PET/SPECT | 3+ | 3+ |
| MRI | 4+ | 3+ |
| Indirect labeling (reporter genes) | ||
| Optical-Fluorescence | 2+ | 3+ |
| Optical-BLI | 2+ | 4+ |
| PET | 3+ | 3+ |
| SPECT | 3+ | 3+ |
| MRI | 4+ | Unknown |
Scale is semi-quantitative: 1+ to 4+ (from least to best spatial resolution/cell detection sensitivity). MRI: magnetic resonance imaging, BLI: bioluminescence imaging, PET: positron emission tomography, SPECT: single photon emission computed tomography. Note these are just qualitative indices and other factors such as depth of signal in optical imaging, pre-loading of cells with imaging agent(s), and exact imaging instrument being used can markedly influence the imaging results.
Similarly, when the goal is to study the biology of these cells and whether they do or do not express certain gene(s) or do or do not come in contact with the environment, the imaging strategy to be used has to be one that will only emit signal or even ceases to, when the biological action being studied is taking place (e.g., differentiation of a stem cell into an adult cell) and a specific pathways is being activated. At the present time, reporter gene imaging appears to provide the best tool to address these questions. For example, if one wants to study whether a stem cell has differentiated into an adult myocyte, use of a reporter gene that is driven by a promoter that will only be activated when the cell has the features of an adult myocyte (i.e., expresses the sarcomeric protein Troponin T) can provide that information. Again, the imaging modality to be used (PET vs SPECT vs. optical) should be decided based on the variables previously mentioned.
Summary
Over the last 2 decades, we have seen a revolution in non-invasive stem cell imaging in the living subject. The uncertain results of different trials (especially in the cardiovascular field) highlight even more the importance of using tools like the ones described. In this review, we have outlined some of the most important characteristics of direct- and indirect- cell labeling focusing on reporter gene technology, which will likely be the preferred methodology for long-term monitoring of stem cell biology. It is likely that not one technique will answer all the questions; but use of a multimodality approach will be the more appropriate approach to address the myriad of aspects posed in this exciting and rapidly evolving field.
CONFLICT OF INTEREST
None.
STEMJOURNAL OPEN REVIEW
The evaluations from reviewers for this article are freely available and can be found as supplementary material here: https://dx.doi.org/10.3233/STJ-190003.
ACKNOWLEDGMENTS
NIH HL119795 (MR-P).
REFERENCES
|
[1] |
Baizabal JM , Furlan-Magaril M , Santa-Olalla J , Covarrubias L . Neural stem cells in development and regenerative medicine. Arch Med Res. 2003;34:572–88. |
|
[2] |
Gimble JM , Katz AJ , Bunnell BA . Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100:1249–60. |
|
[3] |
Goldberg JL , Laughlin MJ , Pompili VJ . Umbilical cord blood stem cells: Implications for cardiovascular regenerative medicine. J Mol Cell Cardiol. 2007;42:912–20. |
|
[4] |
Lechner A . Stem cells and regenerative medicine for the treatment of type 1 diabetes: The challenges lying ahead. Pediatr Diabetes. 2004;5(Suppl 2):88–93. |
|
[5] |
Yamada S , Terada K , Ueno Y , et al. Differentiation of adult hepatic stem-like cells into pancreatic endocrine cells. Cell Transplant. 2005;14:647–53. |
|
[6] |
De Bari C , Dell’Accio F , Tylzanowski P , Luyten FP . Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44:1928–42. |
|
[7] |
Liu Y , Shu XZ , Prestwich GD . Osteochondral defect repair with autologous bone marrow-derived mesenchymal stem cells in an injectable, in situ, cross-linked synthetic extracellular matrix. Tissue Eng. 2006;12:3405–16. |
|
[8] |
Anversa P , Leri A , Kajstura J . Cardiac regeneration. J Am Coll Cardiol. 2006;47:1769–76. |
|
[9] |
Ii M , Nishimura H , Iwakura A , et al. Endothelial progenitor cells are rapidly recruited to myocardium and mediate protective effect of ischemic preconditioning via “imported” nitric oxide synthase activity. Circulation. 2005;111:1114–20. |
|
[10] |
Rafii S , Lyden D . Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nat Med. 2003;9:702–12. |
|
[11] |
Genre D , Viens P , Bertucci F , et al. Modulations of dose intensity of doxorubicin and cyclophosphamide in association with G-CSF and peripheral blood stem cells in adjuvant chemotherapy for breast cancer: Comparative evaluation of completion and safety of three intensive regimens. Bone Marrow Transplant. 2002;29:881–6. |
|
[12] |
Thakur ML , Coleman RE , Mayhall CG , Welch MJ Jr. . Preparation and evaluation of 111In-labeled leukocytes as an abscess imaging agent in dogs. Radiology. 1976;119:731. |
|
[13] |
Thakur ML , Lavender JP , Arnot RN , Silvester DJ , Segal AW . Indium-111-labeled autologous leukocytes in man. J Nucl Med. 1977;18:1014–21. |
|
[14] |
Guzman R , Uchida N , Bliss TM , et al. Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc Natl Acad Sci U S A. 2007;104:10211–6. |
|
[15] |
Rice CM , Halfpenny CA , Scolding NJ . Stem cells for the treatment of neurological disease. Transfus Med. 2003;13:351–61. |
|
[16] |
Chakraborty SK , Fitzpatrick JA , Phillippi JA , et al. Cholera toxin B conjugated quantum dots for live cell labeling. Nano Lett. 2007;7:2618–26. |
|
[17] |
Rosen AB , Kelly DJ , Schuldt AJ , et al. Finding fluorescent needles in the cardiac haystack: Tracking human mesenchymal stem cells labeled with quantum dots for quantitative in vivo three-dimensional fluorescence analysis. Stem Cells. 2007;25:2128–38. |
|
[18] |
Lin S , Xie X , Patel MR , et al. Quantum dot imaging for embryonic stem cells. BMC Biotechnol. 2007;7:67. |
|
[19] |
Keppler A , Arrivoli C , Sironi L , Ellenberg J . Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum. Biotechniques. 2006;41:167–70, 172, 174-5. |
|
[20] |
Voura EB , Jaiswal JK , Mattoussi H , Simon SM . Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat Med. 2004;10:993–8. |
|
[21] |
Askenasy N , Zorina T , Farkas DL , Shalit I . Transplanted hematopoietic cells seed in clusters in recipient bone marrow in vivo. Stem Cells. 2002;20:301–10. |
|
[22] |
Kang WJ , Kang HJ , Kim HS , et al. Tissue distribution of 18F-FDG-labeled peripheral hematopoietic stem cells after intracoronary administration in patients with myocardial infarction. J Nucl Med. 2006;47:1295–301. |
|
[23] |
Rini JN , Bhargava KK , Tronco GG , et al. PET with FDG-labeled leukocytes versus scintigraphy with 111In-oxine-labeled leukocytes for detection of infection. Radiology. 2006;238:978–87. |
|
[24] |
Adonai N , Nguyen KN , Walsh J , et al. Ex vivo cell labeling with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) for imaging cell trafficking in mice with positron-emission tomography. Proc Natl Acad Sci U S A. 2002;99:3030–5. |
|
[25] |
Bindslev L , Haack-Sorensen M , Bisgaard K , et al. Labelling of human mesenchymal stem cells with indium-111 for SPECT imaging: Effect on cell proliferation and differentiation. Eur J Nucl Med Mol Imaging. 2006;33:1171–7. |
|
[26] |
Hill JM , Dick AJ , Raman VK , et al. Serial cardiac magnetic resonance imaging of injected mesenchymal stem cells. Circulation. 2003;108:1009–14. |
|
[27] |
Rogers WJ , Meyer CH , Kramer CM . Technology insight: In vivo cell tracking by use of MRI. Nat Clin Pract Cardiovasc Med. 2006;3:554–62. |
|
[28] |
Michalet X , Pinaud FF , Bentolila LA , et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–44. |
|
[29] |
Kalchenko V , Shivtiel S , Malina V , et al. Use of lipophilic near-infrared dye in whole-body optical imaging of hematopoietic cell homing. J Biomed Opt. 2006;11:050507. |
|
[30] |
Yukawa H , Baba Y . In vivo imaging technology of transplanted stem cells using quantum dots for regenerative medicine. Analytical Sciences. 2018;34:525–32. |
|
[31] |
Seleverstov O , Zabirnyk O , Zscharnack M , et al. Quantum dots for human mesenchymal stem cells labeling. A size-dependent autophagy activation. Nano Lett. 2006;6:2826–32. |
|
[32] |
Shah BS , Clark PA , Moioli EK , Stroscio MA , Mao JJ . Labeling of mesenchymal stem cells by bioconjugated quantum dots. Nano Lett. 2007;7:3071–9. |
|
[33] |
Bengel FM , Schachinger V , Dimmeler S . Cell-based therapies and imaging in cardiology. Eur J Nucl Med Mol Imaging. 2005;32(Suppl 2):S404–16. |
|
[34] |
Massoud TF , Gambhir SS . Molecular imaging in living subjects: Seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–80. |
|
[35] |
Montet X , Figueiredo JL , Alencar H , et al. Tomographic fluorescence imaging of tumor vascular volume in mice. Radiology. 2007;242:751–8. |
|
[36] |
Montet X , Ntziachristos V , Grimm J , Weissleder R . Tomographic fluorescence mapping of tumor targets. Cancer Res. 2005;65:6330–6. |
|
[37] |
Roy R , Thompson AB , Godavarty A , Sevick-Muraca EM . Tomographic fluorescence imaging in tissue phantoms: A novel reconstruction algorithm and imaging geometry. IEEE Trans Med Imaging. 2005;24:137–54. |
|
[38] |
de la Zerda A , Bodapati S , Teed R , et al. A comparison between time domain and spectral imaging systems for imaging quantum dots in small living animals. Mol Imaging Biol. 2010;12:500–8. |
|
[39] |
Rice WL , Shcherbakova DM , Verkhusha VV , Kumar AT . In vivo tomographic imaging of deep-seated cancer using fluorescence lifetime contrast. Cancer Res. 2015;75:1236–43. |
|
[40] |
Himes N , Min JY , Lee R , et al. In vivo MRI of embryonic stem cells in a mouse model of myocardial infarction. Magn Reson Med. 2004;52:1214–9. |
|
[41] |
Arbab AS , Bashaw LA , Miller BR , et al. Intracytoplasmic tagging of cells with ferumoxides and transfection agent for cellular magnetic resonance imaging after cell transplantation: Methods and techniques. Transplantation. 2003;76:1123–30. |
|
[42] |
Frank JA , Miller BR , Arbab AS , et al. Clinically applicable labeling of mammalian and stem cells by combining superparamagnetic iron oxides and transfection agents. Radiology. 2003;228:480–7. |
|
[43] |
Bos C , Delmas Y , Desmouliere A , et al. In vivo MR imaging of intravascularly injected magnetically labeled mesenchymal stem cells in rat kidney and liver. Radiology. 2004;233:781–9. |
|
[44] |
Di Tucci AA , Matta G , Deplano S , et al. Myocardial iron overload assessment by T2* magnetic resonance imaging in adult transfusion dependent patients with acquired anemias. Haematologica. 2008;93:1385–8. |
|
[45] |
Ittrich H , Lange C , Togel F , et al. In vivo magnetic resonance imaging of iron oxide-labeled, arterially-injected mesenchymal stem cells in kidneys of rats with acute ischemic kidney injury: Detection and monitoring at 3T. J Magn Reson Imaging. 2007;25:1179–91. |
|
[46] |
Kraitchman DL , Heldman AW , Atalar E , et al. In vivo magnetic resonance imaging of mesenchymal stem cells in myocardial infarction. Circulation. 2003;107:2290–3. |
|
[47] |
Chen IY , Greve JM , Gheysens O , et al. Comparison of optical bioluminescence reporter gene and superparamagnetic iron oxide MR contrast agent as cell markers for noninvasive imaging of cardiac cell transplantation. Mol Imaging Biol. 2008. |
|
[48] |
Dick AJ , Guttman MA , Raman VK , et al. Magnetic resonance fluoroscopy allows targeted delivery of mesenchymal stem cells to infarct borders in Swine. Circulation. 2003;108:2899–904. |
|
[49] |
Watson DJ , Walton RM , Magnitsky SG , et al. Structure-specific patterns of neural stem cell engraftment after transplantation in the adult mouse brain. Hum Gene Ther. 2006;17:693–704. |
|
[50] |
Kraitchman DL , Tatsumi M , Gilson WD , et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation. 2005;112:1451–61. |
|
[51] |
Castaneda RT , Khurana A , Khan R , Daldrup-Link HE . Labeling stem cells with ferumoxytol, an FDA-approved iron oxide nanoparticle. J Vis Exp. 2011:e3482–e3482. |
|
[52] |
Lee NK , Kim HS , Yoo D , et al. Magnetic resonance imaging of ferumoxytol-labeled human mesenchymal stem cells in the mouse brain. Stem Cell Rev Rep. 2017;13:127–38. |
|
[53] |
Skelton RJP , Khoja S , Almeida S , et al. Magnetic resonance imaging of iron oxide-labeled human embryonic stem cell-derived cardiac progenitors. Stem Cells Trans Med. 2016;5:67–74. |
|
[54] |
Bull E , Madani SY , Sheth R , et al. Stem cell tracking using iron oxide nanoparticles. Int J Nanomedicine. 2014;9:1641–53. |
|
[55] |
Li Z , Suzuki Y , Huang M , et al. Comparison of reporter gene and iron particle labeling for tracking fate of human embryonic stem cells and differentiated endothelial cells in living subjects. Stem Cells. 2008;26:864–73. |
|
[56] |
Thakur ML , Segal AW , Louis L , et al. Indium-111-labeled cellular blood components: Mechanism of labeling and intracellular location in human neutrophils. J Nucl Med. 1977;18:1022–6. |
|
[57] |
Wollert KC , Meyer GP , Lotz J , et al. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: The BOOST randomised controlled clinical trial. Lancet. 2004;364:141–8. |
|
[58] |
Chin BB , Nakamoto Y , Bulte JW , et al. 111In oxine labelled mesenchymal stem cell SPECT after intravenous administration in myocardial infarction. Nucl Med Commun. 2003;24:1149–54. |
|
[59] |
Asiedu KO , Ferdousi M , Ton PT , et al. Bone marrow cell homing to sites of acute tibial fracture: (89)Zr-oxine cell labeling with positron emission tomographic imaging in a mouse model. EJNMMI Res. 2018;8:109. |
|
[60] |
Yang B , Brahmbhatt A , Nieves Torres E , et al. Tracking and therapeutic value of human adipose tissue-derived mesenchymal stem cell transplantation in reducing venous neointimal hyperplasia associated with arteriovenous fistula. Radiology. 2016;279:513–22. |
|
[61] |
Wu JC , Tseng JR , Gambhir SS . Molecular imaging of cardiovascular gene products. J Nucl Cardiol. 2004;11:491–505. |
|
[62] |
Troy T , Jekic-McMullen D , Sambucetti L , Rice B . Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9–23. |
|
[63] |
Carr HM , Smyth JV , Rooney OB , et al. Limitations of in-vitro labeling of endothelial cells with indium-111 oxine. Cell Transplant. 1995;4:291–6. |
|
[64] |
Zanzonico P , Koehne G , Gallardo HF , et al. [131I]FIAU labeling of genetically transduced, tumor-reactive lymphocytes: Cell-level dosimetry and dose-dependent toxicity. Eur J Nucl Med Mol Imaging. 2006;33:988–97. |
|
[65] |
Contag CH , Jenkins D , Contag PR , Negrin RS . Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2:41–52. |
|
[66] |
Herschman HR . Noninvasive imaging of reporter gene expression in living subjects. Adv Cancer Res. 2004;92:29–80. |
|
[67] |
Inubushi M , Tamaki N . Radionuclide reporter gene imaging for cardiac gene therapy. Eur J Nucl Med Mol Imaging. 2007;34(Suppl 1):S27–33. |
|
[68] |
Kang JH , Chung JK . Molecular-genetic imaging based on reporter gene expression. J Nucl Med. 2008;49(Suppl 2):164S–79S. |
|
[69] |
Gambhir SS , Barrio JR , Phelps ME , et al. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci U S A. 1999;96:2333–8. |
|
[70] |
Phelps ME . Inaugural article: Positron emission tomography provides molecular imaging of biological processes. Proc Natl Acad Sci U S A. 2000;97:9226–33. |
|
[71] |
Zhang SJ , Wu JC . Comparison of imaging techniques for tracking cardiac stem cell therapy. J Nucl Med. 2007;48:1916–9. |
|
[72] |
Forss-Petter S , Danielson PE , Catsicas S , et al. Transgenic mice expressing beta-galactosidase in mature neurons under neuron-specific enolase promoter control. Neuron. 1990;5:187–97. |
|
[73] |
Naciff JM , Behbehani MM , Misawa H , Dedman JR . Identification and transgenic analysis of a murine promoter that targets cholinergic neuron expression. J Neurochem. 1999;72:17–28. |
|
[74] |
Himes SR , Shannon MF . Assays for transcriptional activity based on the luciferase reporter gene. Methods Mol Biol. 2000;130:165–74. |
|
[75] |
Zhuo L , Sun B , Zhang CL , et al. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev Biol. 1997;187:36–42. |
|
[76] |
Contag CH , Spilman SD , Contag PR , et al. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–31. |
|
[77] |
Krishnan M , Park JM , Cao F , et al. Effects of epigenetic modulation on reporter gene expression: Implications for stem cell imaging. Faseb J. 2006;20:106–8. |
|
[78] |
Cao F , Wagner RA , Wilson KD , et al. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3:e3474. |
|
[79] |
Wu JC , Cao F , Dutta S , et al. Proteomic analysis of reporter genes for molecular imaging of transplanted embryonic stem cells. Proteomics. 2006;6:6234–49. |
|
[80] |
Wang F , Dennis JE , Awadallah A , et al. Transcriptional profiling of human mesenchymal stem cells transduced with reporter genes for imaging. Physiol Genomics. 2008. |
|
[81] |
Contag CH . In vivo pathology: Seeing with molecular specificity and cellular resolution in the living body. Annu Rev Pathol. 2007;2:277–305. |
|
[82] |
Shah K , Jacobs A , Breakefield XO , Weissleder R . Molecular imaging of gene therapy for cancer. Gene Ther. 2004;11:1175–87. |
|
[83] |
Keren S , Gheysens O , Levin CS , Gambhir SS . A comparison between a time domain and continuous wave small animal optical imaging system. IEEE Trans Med Imaging. 2008;27:58–63. |
|
[84] |
Kumar AT , Raymond SB , Dunn AK , Bacskai BJ , Boas DA . A time domain fluorescence tomography system for small animal imaging. IEEE Trans Med Imaging. 2008;27:1152–63. |
|
[85] |
Model R , Orlt M , Walzel M , Hunlich R . Optical imaging: Three-dimensional approximation and perturbation approaches for time-domain data. Appl Opt. 1998;37:7968–76. |
|
[86] |
Tanenbaum ME , Gilbert LA , Qi LS , Weissman JS , Vale RD . A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. |
|
[87] |
Li Z , Wu JC , Sheikh AY , et al. Differentiation, survival, and function of embryonic stem cell derived endothelial cells for ischemic heart disease. Circulation. 2007;116:I46–54. |
|
[88] |
Shinnawi R , Huber I , Maizels L , et al. Monitoring human-induced pluripotent stem cell-derived cardiomyocytes with genetically encoded calcium and voltage fluorescent reporters. Stem Cell Rep. 2015;5:582–96. |
|
[89] |
Chen IY , Wu JC , Min JJ , et al. Micro-positron emission tomography imaging of cardiac gene expression in rats using bicistronic adenoviral vector-mediated gene delivery. Circulation. 2004;109:1415–20. |
|
[90] |
Wu JC , Chen IY , Sundaresan G , et al. Molecular imaging of cardiac cell transplantation in living animals using optical bioluminescence and positron emission tomography. Circulation. 2003;108:1302–5. |
|
[91] |
Rodriguez-Porcel M , Gheysens O , Chen IY , Wu JC , Gambhir SS . Image-guided cardiac cell delivery using high-resolution small-animal ultrasound. Mol Ther. 2005;12:1142–7. |
|
[92] |
Loening AM , Wu AM , Gambhir SS . Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–3. |
|
[93] |
Loening AM , Fenn TD , Wu AM , Gambhir SS . Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel. 2006;19:391–400. |
|
[94] |
Okada S , Ishii K , Yamane J , et al. In vivo imaging of engrafted neural stem cells: Its application in evaluating the optimal timing of transplantation for spinal cord injury. Faseb J. 2005;19:1839–41. |
|
[95] |
Cao F , Drukker M , Lin S , et al. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Clon Stem Cell. 2007;9:107–17. |
|
[96] |
Cao F , Lin S , Xie X , et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–14. |
|
[97] |
Springer ML , Sievers RE , Viswanathan MN , et al. Closed-chest cell injections into mouse myocardium guided by high-resolution echocardiography. Am J Physiol Heart Circ Physiol. 2005;289:H1307–14. |
|
[98] |
Roth DJ , Jansen ED , Powers AC , Wang TG . A novel method of monitoring response to islet transplantation: Bioluminescent imaging of an NF-kB transgenic mouse model. Transplantation. 2006;81:1185–90. |
|
[99] |
Kutschka I , Chen IY , Kofidis T , et al. Collagen matrices enhance survival of transplanted cardiomyoblasts and contribute to functional improvement of ischemic rat hearts. Circulation. 2006;114:I167–73. |
|
[100] |
Kutschka I , Kofidis T , Chen IY , et al. Adenoviral human BCL-2 transgene expression attenuates early donor cell death after cardiomyoblast transplantation into ischemic rat hearts. Circulation. 2006;114:I174–80. |
|
[101] |
Psaltis PJ , Peterson KM , Xu RD , et al. Noninvasive monitoring of oxidative stress in transplanted mesenchymal stromal cells. JACC-Cardiovasc Imag. 2013;6:795–802. |
|
[102] |
Franchi F , Peterson KM , Paulmurugan R , et al. Noninvasive monitoring of the mitochondrial function in mesenchymal stromal cells. Mol Imaging Biol. 2016;18:510–8. |
|
[103] |
Soloviev VY . Tomographic bioluminescence imaging with varying boundary conditions. Appl Opt. 2007;46:2778–84. |
|
[104] |
Lv Y , Tian J , Cong W , et al. Spectrally resolved bioluminescence tomography with adaptive finite element analysis: Methodology and simulation. Phys Med Biol. 2007;52:4497–512. |
|
[105] |
Chen IY , Gheysens O , Ray S , et al. Indirect imaging of cardiac-specific transgene expression using a bidirectional two-step transcriptional amplification strategy. Gene Ther. 2010;17:827–38. |
|
[106] |
Iyer M , Salazar FB , Lewis X , et al. Non-invasive imaging of a transgenic mouse model using a prostate-specific two-step transcriptional amplification strategy. Transgenic Res. 2005;14:47–55. |
|
[107] |
Iyer M , Salazar FB , Lewis X , et al. Noninvasive imaging of enhanced prostate-specific gene expression using a two-step transcriptional amplification-based lentivirus vector. Mol Ther. 2004;10:545–52. |
|
[108] |
Iyer M , Wu L , Carey M , et al. Two-step transcriptional amplification as a method for imaging reporter gene expression using weak promoters. Proc Natl Acad Sci U S A. 2001;98:14595–600. |
|
[109] |
Adams ST Jr , Miller SC . Beyond D-luciferin: Expanding the scope of bioluminescence imaging in vivo. Curr Opin Chem Biol. 2014;21:112–120. |
|
[110] |
Jones KA , Porterfield WB , Rathbun CM , et al. Orthogonal luciferase–luciferin pairs for bioluminescence imaging. J Am Chem Soc. 2017;139:2351–8. |
|
[111] |
Jurgielewicz P , Harmsen S , Wei E , et al. New imaging probes to track cell fate: reporter genes in stem cell research. Cell Mol Life Sci. 2017;74:4455–69. |
|
[112] |
Tjuvajev JG , Avril N , Oku T , et al. Imaging herpes virus thymidine kinase gene transfer and expression by positron emission tomography. Cancer Res. 1998;58:4333–41. |
|
[113] |
Gambhir SS , Barrio JR , Wu L , et al. Imaging of adenoviral-directed herpes simplex virus type 1 thymidine kinase reporter gene expression in mice with radiolabeled ganciclovir. J Nucl Med. 1998;39:2003–11. |
|
[114] |
Herschman HR . PET reporter genes for noninvasive imaging of gene therapy, cell tracking and transgenic analysis. Crit Rev Oncol Hematol. 2004;51:191–204. |
|
[115] |
Ponomarev V , Doubrovin M , Shavrin A , et al. A human-derived reporter gene for noninvasive imaging in humans: Mitochondrial thymidine kinase type 2. J Nucl Med. 2007;48:819–26. |
|
[116] |
Hsieh CH , Chen FD , Wang HE , et al. Generation of destabilized herpes simplex virus type 1 thymidine kinase as transcription reporter for PET reporter systems in molecular genetic imaging. J Nucl Med. 2008;49:142–50. |
|
[117] |
MacLaren DC , Gambhir SS , Satyamurthy N , et al. Repetitive, non-invasive imaging of the dopamine D2 receptor as a reporter gene in living animals. Gene Ther. 1999;6:785–91. |
|
[118] |
Liang Q , Satyamurthy N , Barrio JR , et al. Noninvasive, quantitative imaging in living animals of a mutant dopamine D2 receptor reporter gene in which ligand binding is uncoupled from signal transduction. Gene Ther. 2001;8:1490–8. |
|
[119] |
Kim SJ , Doudet DJ , Studenov AR , et al. Quantitative micro positron emission tomography (PET) imaging for the in vivo determination of pancreatic islet graft survival. Nat Med. 2006;12:1423–8. |
|
[120] |
Lu Y , Dang H , Middleton B , et al. Long-term monitoring of transplanted islets using positron emission tomography. Mol Ther. 2006;14:851–6. |
|
[121] |
Lu Y , Dang H , Middleton B , et al. Noninvasive imaging of islet grafts using positron-emission tomography. Proc Natl Acad Sci U S A. 2006;103:11294–9. |
|
[122] |
Dwyer RM , Ryan J , Havelin RJ , et al. Mesenchymal stem cell-mediated delivery of the sodium iodide symporter supports radionuclide imaging and treatment of breast cancer. Stem Cell. 2011;29:1149–57. |
|
[123] |
Kang JH , Lee DS , Paeng JC , et al. Development of a sodium/iodide symporter (NIS)-transgenic mouse for imaging of cardiomyocyte-specific reporter gene expression. J Nucl Med. 2005;46:479–83. |
|
[124] |
Kim YH , Lee DS , Kang JH , et al. Reversing the silencing of reporter sodium/iodide symporter transgene for stem cell tracking. J Nucl Med. 2005;46:305–11. |
|
[125] |
Miyagawa M , Anton M , Wagner B , et al. Non-invasive imaging of cardiac transgene expression with PET: comparison of the human sodium/iodide symporter gene and HSV1-tk as the reporter gene. Eur J Nucl Med Mol Imaging. 2005;32:1108–14. |
|
[126] |
Terrovitis J , Kwok KF , Lautamaki R , et al. Ectopic expression of the sodium-iodide symporter enables imaging of transplanted cardiac stem cells in vivo by single-photon emission computed tomography or positron emission tomography. J Am Coll Cardiol. 2008;52:1652–60. |
|
[127] |
Zinn KR , Chaudhuri TR . The type 2 human somatostatin receptor as a platform for reporter gene imaging. Eur J Nucl Med Mol Imaging. 2002;29:388–99. |
|
[128] |
Sharma V , Luker GD , Piwnica-Worms D . Molecular imaging of gene expression and protein function in vivo with PET and SPECT. J Magn Reson Imaging. 2002;16:336–51. |
|
[129] |
Schillaci O . Somatostatin receptor imaging in patients with neuroendocrine tumors: Not only SPECT? J Nucl Med. 2007;48:498–500. |
|
[130] |
Zhernosekov K , Aschoff P , Filosofov D , et al. Visualisation of a somatostatin receptor-expressing tumour with 67Ga-DOTATOC SPECT. Eur J Nucl Med Mol Imaging. 2005;32:1129. |
|
[131] |
Alvarez-Maya I , Navarro-Quiroga I , Meraz-Rios MA , Aceves J , Martinez-Fong D . In vivo gene transfer to dopamine neurons of rat substantia nigra via the high-affinity neurotensin receptor. Mol Med. 2001;7:186–92. |
|
[132] |
Tavares D , Tully K , Dobner PR . Sequences required for induction of neurotensin receptor gene expression during neuronal differentiation of N1E-115 neuroblastoma cells. J Biol Chem. 1999;274:30066–79. |
|
[133] |
Kreuzer J , Denger S , Reifers F , et al. Adenovirus-assisted lipofection: Efficient in vitro gene transfer of luciferase and cytosine deaminase to human smooth muscle cells. Atherosclerosis. 1996;124:49–60. |
|
[134] |
Lee CH , Wu CL , Shiau AL . Hypoxia-induced cytosine deaminase gene expression for cancer therapy. Hum Gene Ther. 2007;18:27–38. |
|
[135] |
Gu E , Chen W-Y , Gu J , Burridge P , Wu JC . Molecular imaging of stem cells: Tracking survival, biodistribution, tumorigenicity, and immunogenicity. Theranostics. 2012;2:335–45. |
|
[136] |
Gonzalez-Gonzalez E , Speaker TJ , Hickerson RP , et al. Silencing of reporter gene expression in skin using siRNAs and expression of plasmid DNA delivered by a soluble protrusion array device (PAD). Mol Ther. 2010;18:1667–74. |
|
[137] |
Watkins C , Lau S , Thistlethwaite R , Hopkins J , Harkiss GD . Analysis of reporter gene expression in ovine dermis and afferent lymph dendritic cells in vitro and in vivo. Vet Immunol Immunopathol. 1999;72:125–33. |
|
[138] |
Yaghoubi SS , Campbell DO , Radu CG , Czernin J . Positron emission tomography reporter genes and reporter probes: Gene and cell therapy applications. Theranostics. 2012;2:374–391. |
|
[139] |
Ray P , Tsien R , Gambhir SS . Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Res. 2007;67:3085–93. |
|
[140] |
Kesarwala AH , Prior JL , Sun J , et al. Second-generation triple reporter for bioluminescence, micro-positron emission tomography, and fluorescence imaging. Mol Imaging. 2006;5:465–74. |
|
[141] |
Cohen B , Dafni H , Meir G , Harmelin A , Neeman M . Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–17. |
|
[142] |
Gilad AA , Winnard PT Jr , van Zijl PC , Bulte JW . Developing MR reporter genes: Promises and pitfalls. NMR Biomed. 2007;20:275–90. |
|
[143] |
Naumova AV , Reinecke H , Yarnykh V , et al. Ferritin overexpression for noninvasive magnetic resonance imaging-based tracking of stem cells transplanted into the heart. Mol Imag. 2010;9:201–10. |
|
[144] |
Weissleder R , Simonova M , Bogdanova A , et al. MR imaging and scintigraphy of gene expression through melanin induction. Radiology. 1997;204:425–9. |
|
[145] |
Bloor CM , White FC , Roth DM . The pig as a model of myocardial ischemia and gradual coronary artery occlusion. In: SwindleM.M., MoodyD.C.. PhillipsL. D., eds. Swine as models in biomedical research Ames, Iowa: Iowa State University Press. 1992, 163–75. |
|
[146] |
Rodriguez-Porcel M , Brinton TJ , Chen IY , et al. Reporter gene imaging following percutaneous delivery in swine moving toward clinical applications. J Am Coll Cardiol. 2008;51:595–7. |
|
[147] |
Miyagawa M , Anton M , Haubner R , et al. PET of cardiac transgene expression: Comparison of 2 approaches based on herpesviral thymidine kinase reporter gene. J Nucl Med. 2004;45:1917–23. |
|
[148] |
Bengel FM , Anton M , Richter T , et al. Noninvasive imaging of transgene expression by use of positron emission tomography in a pig model of myocardial gene transfer. Circulation. 2003;108:2127–33. |
|
[149] |
Willmann JK , Paulmurugan R , Rodriguez-Porcel M , et al. Imaging Gene Expression in Human Mesenchymal Stem Cells: From Small to Large Animals. Radiology. 2009. |
|
[150] |
Yaghoubi SS , Jensen MC , Satyamurthy N , et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6:53–8. |
|
[151] |
Ghodsizad A , Bordel V , Bruckner B , et al. Clinical labeling and imaging of transplanted CD133+/CD34+ stem cells in patients with ischemic heart disease. Heart Surg Forum. 2012;15:E116–18. |
|
[152] |
Li YG , Wei JN , Lu J , Wu XT , Teng GJ . Labeling and tracing of bone marrow mesenchymal stem cells for tendon-to-bone tunnel healing. Knee Surg Sports Traumatol Arthrosc. 2011;19:2153–8. |
|
[153] |
Kassis I , Vaknin-Dembinsky A , Bulte J , Karussis D . Effects of supermagnetic iron oxide labeling on the major functional properties of human mesenchymal stem cells from multiple sclerosis patients. Int J Stem Cells. 2010;3:144–53. |
|
[154] |
Gholamrezanezhad A , Mirpour S , Bagheri M , et al. In vivo tracking of 111In-oxine labeled mesenchymal stem cells following infusion in patients with advanced cirrhosis. Nucl Med Biol. 2011;38:961–7. |
|
[155] |
Caveliers V , De Keulenaer G , Everaert H , et al. In vivo visualization of 111In labeled CD133+ peripheral blood stem cells after intracoronary administration in patients with chronic ischemic heart disease. Q J Nucl Med Mol Imaging. 2007;51:61–6. |
|
[156] |
Cutrera J , Dibra D , Xia X , Li S . Enhancement of reporter gene detection sensitivity by insertion of specific mini-peptide-coding sequences. Cancer Gene Ther. 2010;17:131–40. |
|
[157] |
Karimi M , Goldie LC , Ulgiati D and AbrahamLJ. Integration site-specific transcriptional reporter gene analysis using Flp recombinase targeted cell lines. Biotechniques. 2007;42:217–24. |
|
[158] |
Ochiai H , Sakamoto N , Fujita K , et al. Zinc-finger nuclease-mediated targeted insertion of reporter genes for quantitative imaging of gene expression in sea urchin embryos. Proc Natl Acad Sci U S A. 2012;109:10915–20. |
|
[159] |
Rodriguez-PorcelM, WuJC, and GambhirSS, Molecular imaging of stem cells (July 30, 2009), StemBook, ed. The Stem Cell Research Community, StemBook, doi/10.3824/stembook.1.49.1 |
![]()

