Assessing pluripotency in human cells is inherently an intractable problem. In animal systems, pluripotency can be verified through direct means: pluripotent stem cells can be introduced into an developing embryo and thus the cellular developmental potential of any given in vitro preparation can be directly determined by observing the amount of chimaerism or viability of organisms partially or fully derived from in vitro stem cells (Nagy, Rossant et al., 1993).For obvious ethical reasons such stringent experimentation is impossible in the human system; thus indirect evidence through biomarkers and correlative measures of differentiation potential are the next best level of evidence for human stem cell pluripotency. Many if not most stem cell markers that are widely used for predicting pluripotency in human cells are chosen because of evidence from other species. Yet there are hard limitations to this approach because of two main obstacles: first, using only few markers will erroneously validate potentially abnormal stem cells when these irregular preparations express the limited set of markers similar to normal stem cells. Second, in spite of some striking orthologous similarities, the transfer of mechanistic insight from animals to the human systems for fine-grained determination of pluripotent features is problematic. There are species-specific differences in the cultured pluripotent phenotypes and significant evolutionary structural variations in the transcription network wiring of human and animal cells. We have developed a pragmatic, purely data-driven approach toward empirically defining the “normal” human pluripotent state via interrogation of large scale datasets of genome-wide somatic and pluripotent expression profiles (Müller, Schuldt et al., 2011). Models derived from this dataset can be used to rapidly, confidently and inexpensively assess the pluripotency of human cells through web-based analysis of microarray data from new stem cell preparations. In this chapter, we will introduce the reader into some of the underlying concepts of the tool we call “PluriTest”, demonstrate a real world use case, and discuss the potential as well as the limitations of our approach.
1. Introduction
1.1. Historical and methodological considerations
Stevens and Little discovered in the early 1950’s, that 1–10% of all male129 mice develop spontaneous teratomas depending on experimental and environmental factors (Stevens and Little et al.). They found that these tumors develop into all three germ layers, and also that some tumor cells remain undifferentiated and can give rise to experimental aggressive teratocarcinomas when transplanted into other mice from the same strain.(Stevens et al.) Methods for culturing teratocarcinoma cells were subsequently used in developing the first successful derivation of murine embryonic stem cells (Evans and Kaufman et al., 1981; Martin et al., 1981). In retrospect, Stevens and Little described cancer-initiating stem cells for the first time, and essentially jump-started the evolution of the stem cell field and the development of the refined culture methodologies and analysis paradigms we use today.
Unfortunately, for testing the potency of human pluripotent stem cells, we still haven't conceptually advanced beyond the in vivo teratoma paradigm developed in 60 years ago; the teratoma assay is still regarded by many researchers as the “gold standard” for the assessment of pluripotency.
1.2. Quality standards for assessment of iPSC in the field
This section is intended to provide a succinct overview of several assays for the quality control of human pluripotent stem cells and is neither comprehensive nor complete. No single example of these tests for pluripotency can be regarded as definitive on its own and we suggest that multiple methods can be used to integrate the various kinds of evidence into a comprehensive “life cycle” picture of each hPSC line (Fig. 1).
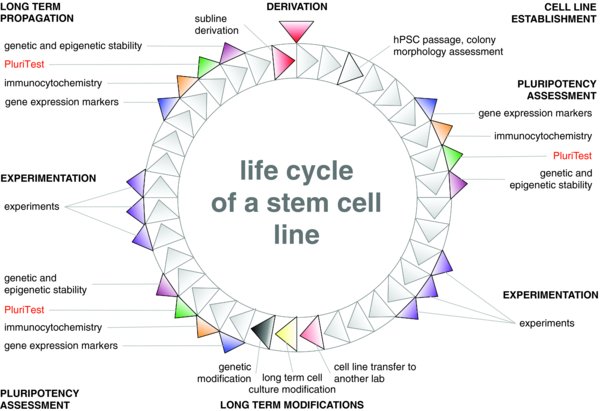
An hPSC line progresses through general stages throughout its life cycle: establishment, experimentation, long-term modification and long-term propagation. An hPSC line can “fork” into different paths. For example, clonal lines and reporter lines have been derived from the WA09 line (e.g. WA09.2). At each of the illustrated steps, a thorough pluripotency assessment is recommended.
Our primary objective is to replace the teratoma assay, but PluriTest could replace other assays as well. We propose an alternative to the teratoma assay, which is expensive and requires animal surgery, using a purely data-driven standardized bioinformatic analysis of genome-wide transcriptional profiles. We provide background information, step-by-step instructions and exemplary interpretations for PluriTest as a prototypic example of such an assay.
The pluripotent stem cell field is expanding rapidly, and surrogate assays for assessing pluripotency, such as PluriTest, are critical for preclinical and translational stem cell research for the foreseeable future. However, proxy assays that rely on a defined set of biomarkers, such as RTPCR panels, immunocytochemistry markers, and methylation status of specific genes, lack the specificity required for selecting only truly pluripotent stem cells (Williams, Schuldt et al., 2011). For example, a positive result using immunocytochemistry or RTPCR for pluripotency-associated genes indicates that pluripotent cells are present, but provides no information about the homogeneity of the population; even 14-day old embryoid bodies are identified as pluripotent by these assays. Similarly, assessment of the methylation status of the promoter of a pluripotency-associated gene would indicate pluripotency for some cancer cells, partially reprogrammed cells, and karyotypically abnormal cells. PluriTest avoids these pitfalls by comparing the whole genome expression profile of the query sample with a model of confirmed undifferentiated euploid hPSCs built from a very large dataset gene expression information (Williams, Schuldt et al., 2011). This assay not only identifies samples as containing pluripotent cells, but also reveals contamination by differentiated cells, and genomic and epigenomic abnormalities that affect gene expression patterns.
PluriTest is a tool for rapid assessment of pluripotency in human in vitro stem cell preparations. We expect that researchers will initially screen for promising stem cell colonies after derivation or experimental manipulation by using RTPCR or immunocytochemistry for detecting pluripotency-associated markers before confirming pluripotency with whole genome expression analysis for PluriTest (Fig. 1). However, with near-perfect sensitivity and specificity, PluriTest can function as the sole means to determine pluripotency in any given culture. We envision that further developments of PluriTest platform will have utility for not only testing for pluripotency, but also identifying differentiation propensity and genomic integrity in stem cell preparations.
1.2.1. Cell culture morphology and dynamics
hESCs and hiPSCs show a distinct colony morphology when cultured on murine embryonic fibroblasts (Thomson, Itskovitz-Eldor et al., 1998; Takahashi, Tanabe et al., 2007). This characteristic feature allows for the identification of successfully reprogrammed iPSCs (Meissner, Wernig et al., 2007) (Fig. 2).

Selecting “good” colonies from “bad” ones is probably the most commonly used quality control step in hPSC research. Here we show iPSCs generated from fibroblasts with episomal vectors and stages of propagating them until the cells have lost the episomes at about passage 15 (Okita, Matsumura et al., 2011). Only colonies with “good” morphology (flat appearance, well defined edges, homogenous cell morphology) are selected in each passage and carried forward.
Visual inspection and manual selection of “good” from “ugly” looking colonies probably remains the most under-appreciated yet most important control instrument for pluripotent quality assessment, used every day in hPSC labs worldwide. Development of the expertise to decide which colonies to pick and which to discard requires apprenticeship with experienced researchers and is difficult to operationalize (Loring, Schwartz et al., 2007). Typical hPSC morphological characteristics are often lost or altered in modern, automatable feeder-free and defined culture conditions (Ludwig, Bergendahl et al., 2006; Wang, Schulz et al., 2007); this is one of the reasons that most preclinical research labs routinely culture and derive hPSCs on feeder cell systems.
1.2.2. Karyotypic and genomic stability
hPSCs need to be karyotyped on a regular basis (Loring, Schwartz et al., 2007). Mounting evidence shows that PSC regularly acquire a pluripotency-specific set of karyotypic abnormalities (Mayshar, Ben-David et al., 2010; Laurent, Ulitsky et al., 2011). Unfortunately, subchromosomal mutations frequently occur in the same genomic regions that are found to be amplified on the karyotypic level (Laurent, Ulitsky et al., 2011). Such small abnormalities can only be detected with genome wide, high resolution SNP (single nucleotide polymorphism) and CGH (comparative genome hybridization) microarrays, or DNA sequencing. Recent evidence suggests that when compared to their respective parental cells, some iPSC lines contain 1000 to 2000 single allele base changes, including more than 10 non-synonymous changes in coding genes(Cheng, Hansen et al., 2012). These data highlight the dynamic character inherent to the life cycle of a hPSC line.
1.2.3. Marker expression (RT-PCR, IHC, cell surface markers)
Almost every manuscript that reports the derivation of hESC and iPSC lines shows immunocytochemical proof of expression of pluripotency-associated markers such as POU5F1 (OCT4), SOX2, NANOG, and/or the antigens TRA1-61, TRA1-81, and SSEA4. Several companies offer “stem cell characterization kits” with these antibody panels. Other commercial assays are based on expression measurements for a group of pluripotency-associated genes. The International Stem Cell Initiative proposed a set of 96 QRT-PCR markers for the assessment of pluripotency and differentiation in hESC lines (Adewumi, Aflatoonian et al., 2007). Commercialized versions of these stem cell biomarker sets are able to positively identify cells as hPSCs, but cannot reliably discriminate aberrant phenotypes such as partially reprogrammed iPSCs, epigenetically or genetically abnormal iPSCs and hESCs, or teratocarcinoma cell lines.
There is a recent trend toward using cell surface markers, such as antibodies (Lowry, Richter et al., 2008; Chan, Ratanasirintrawoot et al., 2009) and lectins (Wang, Nakagawa et al., 2011) that selectively bind to hESCs and iPSCs. The advantage of these markers is that they can be used to sort cells by flow cytometry or magnetic bead-based technology. The magnetic bead technology may become an important technological development to support fully automated high throughput hiPSC derivation and culture methods, and it has already been integrated into improved protocols for automated hiPSC selection (Dick, Matsa et al., 2011; Valamehr, Abujarour et al., 2012) and hPSC passaging.
1.2.4. In vitro differentiation
Numerous in vitro differentiation protocols for all three germ layer lineages and many terminally differentiated somatic cell types have been reported. However, there is no current standard of a single set of in vitro differentiation protocols necessary or sufficient to demonstrate pluripotent differentiation potential. Undirected differentiation into all three germ layers is often performed using an embryoid body protocol (Martin and Evans et al., 1975), in which the hPSCs are aggregated and cultured in suspension until the cells begin to differentiate. There is no common protocol for generating EBs, and no commonly agreed-upon set of markers for quantitatively analyzing the outcome of such an experiment. Even with the comprehensive ISCI 96 stem cell marker set, it has been difficult to reliably discern EBs from undifferentiated hESC cultures, possibly due to residual undifferentiated PSC even in late EB stages (Adewumi, Aflatoonian et al., 2007). Attempts have been made to standardize EB formation (Burridge, Anderson et al., 2007; Pick, Azzola et al., 2007; Ungrin, Joshi et al., 2008), but no commonly accepted standard has emerged so far.
1.2.5. In vivo differentiation (teratoma assay)
The teratoma assay for in vivo demonstration of pluripotential differentiation capabilities based on work by Stevens and Little in the 1950s and 1960s (Stevens and Little et al., 1954; Stevens et al., 1964) was employed in the first-of-its-kind manuscripts demonstrating the derivation of murine and human ESC lines (Evans and Kaufman et al., 1981; Martin et al., 1981; Thomson, Itskovitz-Eldor et al., 1998) as well as murine and human iPSC lines (Takahashi and Yamanaka et al., 2006; Takahashi, Tanabe et al., 2007; Yu, Vodyanik et al., 2007). The assay requires the sacrifice of laboratory animals and is expensive and time consuming (Peterson, Tran et al., 2011).
Although the teratoma assay is regarded by some researchers as still essential for the qualification of new human pluripotent cell lines (Dolgin et al., 2010), in practice it is actually performed for less than half of the new hESC or hiPSC lines reported. In a recent study, we found that from 1998 to 2009 only 44% of 639 hESC lines and only 36% of 777 iPSC lines were actually tested in vivo by the teratoma assay(Müller, Goldmann et al., 2010).
For producing teratomas, usually about on million hPSC are in injected into each of three immunodeficient male mice. Common injection sites are under the skin, into the hind limb muscles, into the kidney capsule, into the testes; one group transplants cells to the spinal cord (Israel, Yuan et al., 2012). The animals need to be monitored on a daily basis for two to three months and their greatest potential for suffering is at the end of that time period, when teratomas have significantly increased in size often due to fluid-filled cysts, with potential for compression of internal organs (Peterson, Tran et al., 2011) and sometimes resulting in ulceration in tumors formed by subcutaneous and intramuscular injection.
The analysis of teratomas is not straightforward and requires the expertise of a trained pathologist.(Gertow, Cedervall et al., 2011) Often, the most benign type of teratomas (called mature or cystic) turns out the least informative. Growth of large fluid-filled cysts suppresses the development of the surrounding stem cell-derived tissues, due to pressure atrophy, resulting in teratomas without discernible differentiation of some germ layer derivatives. Analysis of cellular differentiation in teratomas is usually superficial and qualitative, with most researchers only highlighting islands of identifiable differentiation based on histological features, such as clusters of neural rosettes or secretory glandular tissue.(Müller, Goldmann et al., 2010) In our experience, 30 to 80% of the observable teratoma tissue cannot be confidently assigned to a specific germ layer with conventional histological methods .
Most mature lineages cannot be confidently modeled and observed within teratomas. For example, neural derivatives usually do not progress beyond an immature embryonic or fetal state. Consequently, for disease modeling applications or drug screening efforts requiring mature differentiated cell products such as spinal cord motoneurons(Dimos, Rodolfa et al., 2008) or differentiated cells from body surfaces such as epidermal keratinocytes, the teratoma assay has no informative value for the propensity of an iPSC line to differentiate into these cell types.
1.2.6. Genome wide profiling
Genome wide profiles of pluripotent stem cells were published for the first time in 2001 (Loring, Porter et al., 2001). Since then, profiling of stem cell transcriptomes has become a commonly used methodology for the analysis of newly established hPSC lines to compare them with existing hESC or iPSC lines. In regard to global profiling methods, we propose the distinction of functional versus structural assays. Transcriptomics currently is probably the prime example for a genome wide functional assay. As a highly integrated molecular machinery in each cell is necessary for transcribing snippets of DNA in a genomic locus into various kinds of RNA (mRNA, microRNA and nclRNA), transcriptional profiles are the consequence of an active and highly orchestrated process within each cell. In contrast, epigenomic profiles are not well understood and the data generated by genome wide profiling, of CpG marks for example, cannot yet be directly linked with a relevant cellular function. A gene that has been found to be over-expressed in one hiPSC line and hypothetically connected to a specific function can be easily tested further through over-expression or knockdown of the same gene in several hPSC lines. The same is currently impossible for differentially methylated regions found in one hPSC line when compared to others. Eventually, epigenetic signals will surely be linked with processes governed by and governing cellular states, but currently our knowledge of the epigenetic makeup of stem and somatic cells assays can be interpreted only as structural variations without immediate and obvious insights or easily testable hypotheses.
While it is in principle possible to establish pluripotency assays with all types of genome-wide data such as mirRNA(Laurent, Chen et al., 2008), mRNA(Müller, Laurent et al., 2008; Müller, Schuldt et al., 2011), chromatin modifications (Ernst, Kheradpour et al., 2011) and CpG methylation (Laurent, Wong et al., 2010; Bock, Kiskinis et al., 2011; Nazor, Altun et al., 2012 (in press)), the only currently biologically interpretable signatures are those based on mRNA transcriptomes.
2. Protocols
Here we describe step-by-step how the pluripotency of hPSC samples can be assessed through the PluriTest online tool. With PluriTest, it is straightforward to assess a sample for pluripotency, based on comparison with the gene expression profiles of a large database of cell samples known to be pluripotent.
First, the PluriTest user generates stem cell samples that need to be qualified for pluripotency.
Second, the researcher submits a cell pellet or extracted RNA to a microarray core, which in turn analyzes the cells, perferably on an Illumina HT 12v4 gene expression microarray.
Third, the user retrieves the microarray data from their core facility and uploads the raw data to a free public website at www.pluritest.org (see Fig. 3 for an overview of the online workflow). There, the data is analyzed online with the published PluriTest algorithm(Müller, Schuldt et al., 2011) and the cells are analyzed with two related parameters. These parameters have a distinct interpretation, for which we will provide here in scenario-based examples. For more details on the conceptual background, please refer to Williams, et al., 2011(Müller Schuldt et al., 2011; Williams, Schuldt et al., 2011).

For providing a fair comparison we assumed that for a single hPSC cell line that RNA is run in biological triplicates, with the exception for the NanoString assay, where we assumed that RNA is run in biological duplicates. As we explain in the main text, we use PluriTest regularly only as single point assay or in biological duplicates. For the teratoma assay, we used three mice injected with cells from a cell line as basis for our cost calculation. We have inquired and estimated the costs associated with running an assay in an academic setting and based this calculation on the material costs without factoring labor or equipment costs in. Commercial prices are based on quotes we inquired in California, USA.
We also provide base figures that can help estimating the data depth offered by each assay next to each time/cost point.
2.1. Use cases
Two general scenarios represent the most frequent use cases, pluripotency determination and differentiation assessment.
2.1.1. Pluripotency determination
In the first scenario, a researcher has derived a new iPSC line from a donor or used a novel method for inducing pluripotency in somatic cells. As a result, the researcher has established stable lines that now need to be qualified as pluripotent to be carried further into functional assays, such as differentiation experiments. In this case a stably propagating stem cell line exists, from which a sufficient number of cells can be harvested for multiple assays. PluriTest was designed to offer an economical, time-saving and animal sparing alternative to the teratoma assay, and requires less of the same starting material as needed for teratoma assays (Peterson, Tran et al., 2011) In this scenario, only samples passing the Pluripotency Score and Novelty Score thresholds should be used for experiments that rely on assuredly pluripotent cells.
2.1.2. Differentiation assessment
Another scenario is the use of PluriTest for the assessment of differentiation in cultures. The Novelty Score is highly sensitive to hPSC cultures contaminated with differentiated cells or populations exiting from pluripotency in a differentiation time course. The current version of PluriTest allows a researcher to recognize mixtures of undifferentiated and differentiated cells, but not to identify the specific type of differentiation occurring. To make the assay useful for identifying specific cell types, we are currently establishing a large database of gene expression profiles of multiple well-characterized specifically differentiated cell types and experimental mixtures of undifferentiated and differentiated cells. We envision future versions of PluriTest that enable determination of the direction of differentiation, and position of cells in a global gene expression body map, as well as for unmixing different emerging populations from heterogenous cell cultures.
In its current version PluriTest can determine if a subset of cells in a culture have differentiated. In mixing experiments, we found that adding 25% of human fibroblast RNA to 75% hESC RNA pushes both Pluripotency and Novelty scores outside the empirically set thresholds for hPSC (Müller, Schuldt et al., 2011). However, the scores remain very high, indicative of the large portion of undifferentiated cells (Müller, Schuldt et al., 2011). In differentiation time course experiments, differentiation becomes observable first in the Novelty Score, as this parameter is more sensitive to the acquisition of non-pluripotent signatures within a hPSC culture (Müller, Schuldt et al., 2011).
2.2. Biological samples
PluriTest in its current form requires only one sample of a cell line to globally assess the pluripotent expression profile in a given stem cell line at important steps of a cell line's life cycle (Fig. 1). Ideal candidates for PluriTest are newly established hESC or iPSC lines, cell lines after genetic manipulation (e.g. after correction of a patient cell lines with a genetic defect with TALE nucleases) or cell culture experiments (e.g. siRNA-based knockdown of genes). However, we usually run two biological replicates of a hPSC line in parallel to be sure that we can detect biological or technical outliers.
We advise that the samples and RNA be of the highest possible quality. For assurance of high quality samples, select only the best, undifferentiated cultures and colonies. In our experience, the most important and constant factor for assuring high-quality RNA has been the number of cells used for RNA extraction (ideally >1 million cells). We have successfully used both Life Technology's mirVana and Qiagen's RNA isolation kits. We qualify the RNA with strict quality control criteria, using Life Technology's QBit, Agilent's BioAnalyzer and the NanoDrop device in parallel in our lab.
2.3. Obtaining microarray data
PluriTest currently only supports microarray data from the Illumina HT12v3 and v4 platforms. We advise that researchers not experienced with hands-on microarray analysis should use core facilities. Alternatively, several commercial service providers allow for technically excellent Illumina HT12v4 analyses on a relatively small fee-for-service basis. We recommend that researchers use Illumina-certified service providers ( http://www.illumina.com/services/cspro.ilmn). The PluriTest model has been modified to exclude technical variations stemming from different core services if standard Illumina sample handling, preparation and hybridization protocols are followed (Müller, Schuldt et al., 2011).
Because data analysis was challenging during the early phases of microarray development in the 1990's (long before the human genome was sequenced), there is still a widely prevalent perception that microarray analysis is expensive and difficult. After 15 years of improvements, obtaining microarray data has become straightforward and relatively inexpensive.
The Illumina HT12 microarray platform supported by PluriTest is widely available in core facilities and from service providers. In the Loring lab at the Scripps Research Institute, which has all of the equipment and expertise to run HT12 arrays in house, the cost per sample is approximately US$100-150 for the complete workflow, including RNA extraction, QC array hybridization and analysis. The Müller lab in Kiel has access to fee-per-service microarray analysis at a central facility of the Christian-Albrechts Universität zu Kiel but has outsourced the microarray work for PluriTest mainly to a biotech company located in Denmark for the price of ∼€250 per HT12 array for the complete PluriTest workflow from cell pellet to data download.
These three options, array preparation in lab, at a institution's central array core and outsourced, make it possible for almost all labs world-wide to obtain PluriTest analyses for their hPSC lines. While the outsourcing option is the most expensive, our experience with it has been excellent, as the effort for labor, sample tracking through the analysis and sample process is reduced to a minimum. A list of Illumina certified service providers can be obtained at http://www.illumina.com/services/cspro.ilmn.
Time is an important factor and the teratoma assay usually requires about 2--3 months to completed (Peterson, Tran et al., 2011). In contrast, mRNA expression analysis methods like microarrays (Müller, Schuldt et al., 2011), RT-PCR (Adewumi, Aflatoonian et al., 2007) or NanoString (Bock, Kiskinis et al., 2011) can be usually completed within a single week. Fig. 3 gives an overview of several methodologies that have been proposed for assessment of pluripotency and compares the different assays with regard to time to endpoint and cost.

First microarray data are generated from cell lines, then uploaded to the PluriTest website. There the published PluriTest algorithm and model (marked in red) is used to assess the individual Pluripotency Scores and Novelty Scores for the uploaded samples and reports these back to the user (marked blue).
2.4. Upload of raw array data
It is important to obtain the raw data from the microarray core, as PluriTest uses the most basic, binary file type output from Illumina HT12 arrays (so called .idat files). These .idat files are then uploaded through the website, www.pluritest.org (see Fig. 4).
For this the user generates first a personal account with a password and login name. Then the user logs on and selects the .idat files for upload on the local computer. Currently, the Silverlight 4 or 5 plugin needs to be installed on the local machine in order to access and operate the online PluriTest tool.
2.5. Use of the PluriTest R script
We have published the PluriTest model along with an R script that can be downloaded [ http://www.nature.com/nmeth/journal/v8/n4/full/nmeth.1580.html] The BioConductor script is intended for researchers proficient in bioinformatic programming in R and to enable closer examination of the underlying model and mathematics behind PluriTest. The online PluriTest application uses the same script, and handles all the tasks, which require command line execution of code through an web based conventional point and click interface.
2.6. Interpretation of the Results
After the raw data has been uploaded, the samples analyzed on the PluriTest server and the results are reported back to the user through two related parameters, Pluripotency Score and Novelty Score (as depicted in Fig. 5A).
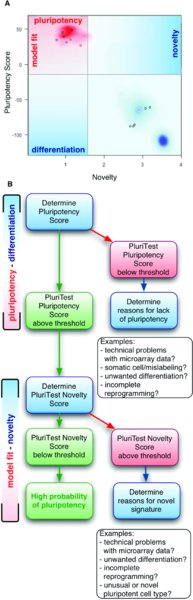
The Pluripotency Score informs the user on how strongly a model-based pluripotency signature is expressed in the samples analyzed. The Novelty Score indicates the general model fit for a given sample: if it is low, the analyzed sample is well represented in the current PluriTest data model, if the Novelty Score is high and the sample also has a high Pluripotency Score, further functional assays might be necessary determine the identity of the sample.
Pluripotency Score: A score that is based on all samples (pluripotent cells, somatic cells and tissues) in a large collection of human pluripotent and somatic cells, which we have termed Stem Cell Matrix (Müller, Laurent et al., 2008; Müller, Schuldt et al., 2011). Samples with high positive Pluripotency Scores are more similar to the pluripotent samples in the model matrix than to all other classes of samples in the matrix (Fig. 5A).
The Pluripotency Score gives an indication if a sample contains a pluripotent signature, but not necessarily if the cell preparation is a normal, bona-fide hESC or hiPSC. Partially differentiated pluripotent cells, teratocarcinoma cells or karyotypically abnormal embryonic stem cells may also have a high Pluripotency Score. For a more detailed explanation of the Pluripotency Score please refer to (Müller, Schuldt et al., 2011; Williams, Schuldt et al., 2011).
Novelty Score: A score that is based on well-characterized pluripotent samples in the stem cell model matrix. Samples with high Novelty Scores are more dissimilar to the pluripotent samples in the model matrix than the other pluripotent samples in the matrix. A low Novelty Score indicates that the test sample can be well reconstructed based on existing data from other well-characterized iPSC and ESC lines (“good model fit”, Fig. 5A). A high Novelty Score indicates that there are patterns in the tested sample that cannot be explained by currently existing data from well-characterized, karyotypic normal pluripotent stem cells. Partially differentiated pluripotent cells, teratocarcinoma cells or karyotypically abnormal embryonic stem cells may have a high pluripotency score but cannot be reconstructed well with data from well-characterized, normal pluripotent stem cells and thus are expected have a high Novelty Score. For a more detailed explanation of the concepts behind the Novelty Score please refer to (Müller, Schuldt et al., 2011; Williams, Schuldt et al., 2011).
PluriTest classifies cells as pluripotent based on empirically determined thresholds. This classification is reported back to the user. The Pluripotency Score and Novelty Score are related, yet non-identical measures for pluripotency in human cells, and we suggest that the outcomes be interpreted in a sequential manner as depicted in Fig. 5B. First the researcher should assess the Pluripotency Score. High values indicate expression of pluripotency-associated genes and lack of expression of differentiation-associated genes in a given sample. Samples in which cells express POU5F1/OCT4, NANOG and KLF4 should usually reach our empirically set thresholds. However, the Pluripotency Score will not identify samples as pluripotent if they are simply overexpressing reprogramming factors (such as in fibroblasts early after transduction and before transition to the pluripotent state), because several thousand genes are used for this assessment. For example, “partially reprogrammed” cell lines (Chan, Ratanasirintrawoot et al., 2009) do not show Pluripotency Score levels comparable to fully reprogrammed iPSC. This feature of PluriTest contrasts with the limited gene sets (such as the ISCI marker panel (Adewumi, Aflatoonian et al., 2007)) assayed by RT-PCR.
The Novelty Score adds information on the “purity’ of the pluripotent signal. It will increase if signatures are present that are not present in our large training dataset of several hundred proven pluripotent hPSC samples. For example, genomically abnormal hESC lines such as the triploid BG01v line (Zeng, Chen et al., 2004) or teratocarcinoma cell lines will be distinguished by unusually high Novelty Scores. Currently, the Novelty Score is set to detect verified genomic and epigenetic abnormalities, so small epigenetic or genetic abnormalities in the pluripotent genomewide transcriptome may be undetected. We are developing improved methods for reliably identifying and scoring reported recurrent small deviations, and we will update the online PluriTest application as soon as the algorithms have been sufficiently validated.
Researchers using PluriTest should be aware that the assay with its empirically determined thresholds is very specific and sensitive for hPSC cells (Müller, Schuldt et al., 2011). The thresholds we have set so far have only resulted in false negatives, i.e. pluripotent cell lines that are capable of teratoma formation and have high Pluripotency Scores, but score above the pluripotency threshold on the Novelty axis. A prototypical example for such a case is parthenote-derived hESCs, which display significant epigenetic differences from conventional hESCs (Harness, Turovets et al., 2011) and are picked up by PluriTest as unusually high scoring samples on the Novelty axis (Müller, Schuldt et al., 2011). Thus, PluriTest can highlight unusual hPSC samples with a hPSC-typical Pluripotency Score and a unusual high Novelty Score.
2.7. Reporting PluriTest results
Finally, when the user receives the PluriTest's assessment of the Pluripotency Score and Novelty Score in their samples they can copy and paste the results into a manuscript that describes, for example, derivation of a set of novel iPSC lines. The 2D PluriTest plot depicted in Fig. 5A and Fig. 6 is the best way to communicate the combined results and put them into the context of the empirical distribution of pluripotent and somatic samples used to construct and validate PluriTest. The background is a density distribution of the Stem Cell Matrix samples (Müller, Schuldt et al., 2011), with the red and blue clouds representing the empirical density distributions for hPSCs and differentiated cells, respectively.
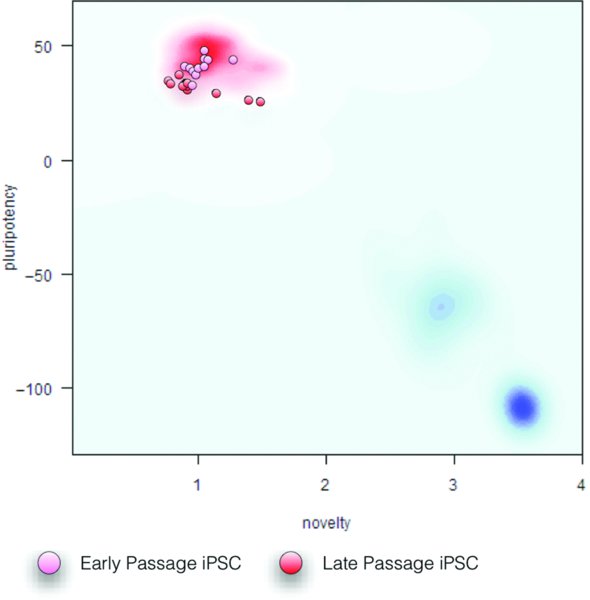
Red and blue clouds in the background represent the empirical density distributions for hPSCs and differentiated cells in the Stem Cell Matrix, respectively. The early and late passage iPSC samples are all pluripotent as determined by their Pluripotency and Novelty scores.
2.8. Getting the PluriTest results published
We have now successfully passed peer review for several manuscripts in which iPSC lines were established for which we have used only PluriTest and not the teratoma assay for validation. We still have received several critiques by referees and have copied one of our answers in the troubleshooting section.
3. Troubleshooting
Problem: The array and PluriTest did not work that well. The RNA input was really low as we are sorting a rare cell type from tissues and/or from cell cultures. The cells did not even give us a QC on the Bioanalyzer pico chip although the cell number input per sample was 12,000 cells.
We still went ahead with the array but just a few samples gave signals over background and seem to cluster together based on cell number input.
Solution: PluriTest was established for the routine testing of human PSC lines with abundant and high-quality starting material. We recommend to beginning with at least 1×106 cells as starting material and to discard all RNA extracted samples with less than perfect RNA QC measures (e.g. the Agilent Bioanalyzer RIN should be always >9). PluriTest has been designed to replace and improve on the teratoma assay, in which about a million cells are used for testing for pluripotency. Any other use is highly experimental and will go beyond PluriTest's validated bounds, yet might still be able to yield interesting results. It is possible for PluriTest to be used in less than perfect settings and with suboptimal materials and still yield excellent outcomes, but we caution against an over-interpretation of these results, particularly if the results indicate that a hPSC sample doesn't fall within the empirically set thresholds for pluripotent cells on the Pluripotency and Novelty Scores.
Problem: We have now successfully run Illumina arrays with our samples and run PluriTest, but the combined Pluripotency and Novelty Scores plot is are hard to read and we would like to add some labels to the plot. What should we do?
Solution: For this purpose, we copy-paste the plot into a graphic editor (such as Omnigraffle or Adobe Illustrator) and label the samples in the plot by hand. For example Fig. 5 was enhanced this way in Omnigraffle.
Problem: We have submitted a manuscript and the reviewers didn't like at all that we used PluriTest instead of the teratoma assay.
Solution: It is important to keep in mind that less than half of the hESC and iPSC lines generated prior to the publication of PluriTest were validated by the teratoma assay (Müller, Goldmann et al., 2010). While this information may be helpful to a reviewer, it may not be convincing; there is a large gap between what researchers/referees believe the current state of the art of pluripotency qualification is, and what most researchers actually do. We have now successfully passed peer review and published studies that have used PluriTest instead of a teratoma assay (Petit, Kesner et al.) and these may be cited. We will share a typical negative criticism for using PluriTest instead of the teratoma assay from a peer review of a high profile study. We provide arguments that can be tried in this situation and hope it may help others with similar problems to come up with a successful rebuttal.
-
Reviewer 1:
-
The authors use PluriTest, which is a transcriptional profile, to claim that these cells are iPS cells. The authors should perform teratoma assays. These are routinely done to demonstrate that cells are indeed “iPS”.
-
-
Response:
-
Several techniques are commonly used to demonstrate the pluripotency of ES or iPS cell lines: alkaline phosphatase activity or expression of pluripotency-associated markers by immunocytochemistry (OCT4, SOX2, TRA-1), production of embryoid bodies (EB) coupled with detection of gene expression associated with the three germ layers, and teratoma formation in immunodeficient mice. In the submitted manuscript, we have used the two first techniques, showing that the wild type and “dipsydoodle-disrupted”- iPSC lines express high levels of pluripotency-specific genes and are able to produce EBs that express specific genes of the three germ layers.
-
We did not perform the teratoma assay for the following reasons: an increasing number of leading labs working on iPSCs have raised concerns about the assay's effectiveness as a stem cell quality control tool. Other researchers have also found that partially reprogrammed iPSCs can form teratomas even if they do not meet other criteria for pluripotency, leading many to question the overall significance of the teratoma assay (E. Dolgin, “Putting stem cells to the test,” Nature Medicine, vol.16, no. 12, pp. 1354–1357, 2010; Müller, F.-J. et al., A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell 6, 412–414 (2010) as well as Chan, E. et al., Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol (2009)).
-
Several recent studies about iPSC and hESC derivation have been published without teratoma data; among them are high profile publications such as the first report of derivation of iPSC from ALS patients published in Science in 2008 (Dimos et al., 2008).
-
In a recent peer-reviewed study we found that of 639 human iPSC lines reported in 95 studies from 2007 to 2009, only 44% were tested with the teratoma assay.(Müller et al., 2010). Also, only 36% of the 777 hESC lines reported in publications from 1998 to 2009 were assayed for their teratoma-forming ability (Müller et al., 2010).
-
Because of its lack of specificity and non-uniform usage, the teratoma assay has become a troublesome approach, particularly for researchers at institutions requiring and enforcing strict ethical use of tumor forming animal experiments.
-
Several techniques have been developed in the last few years to avoid the use of animals unnecessarily for studying tumor formation. One of them, PluritTest (Müller et al., 2011), uses a pattern recognition algorithm that is able to distinguish between pluripotent and non-pluripotent cell lines with near perfect discriminatory power; the only misclassified samples are false negatives caused by technical problems with microarray preparation and hybridization. It relies on a large database of gene expression patterns from known human stem cell lines and has been validated on more than 6000 microarray assays run in many labs worldwide (Müller et al., 2011; plus > 2500 user data sets from the PluriTest website as of March, 2012). The main advantage of the PluriTest over the teratoma assay is that it doesn't require animal experimentation to determine if a specific cell line is pluripotent. An additional feature of the PluriTest diagnostic test is that it can detect unexpected variation in the pluripotent phenotype on a global scale, which the teratoma assay in its usual use does not. It is important to note that teratocarcinoma cell lines, genetically and/or epigenetically abnormal hPSCs may be able to form teratomas; the teratoma assay was originally developed using serially transplanted murine teratocarcinomas.(Stevens & Little 1954; Stevens 1964). We expect that PluriTest-like global transcriptome and/or epigenetic assays to become an important tool to complement SNP genotyping and karyotyping to detect abnormalities in hPSC lines.
-
Dimos, J.T. et al., 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science (New York, NY), 321(5893), pp.1218–1221.
-
Müller, F.-J. et al., 2011. A bioinformatic assay for pluripotency in human cells. Nature methods.
-
Müller, F.-J. et al., 2010. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell, 6(5), pp.412–414.
-
Stevens, L.C., 1964. Experimental production of testicular teratomas in mice. Proceedings of the National Academy of Sciences of the United States of America, 52, pp.654–661.
-
Stevens, L.C. & Little, C.C., 1954. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proceedings of the National Academy of Sciences of the United States of America, 40(11), pp.1080–1087.
-
As a response to our rebuttal (and acceptance of our manuscript), the reviewer wrote the following:
-
We appreciate the authors’ extensive comments on the Pluritest. […] For human iPS cells, it seems that the Pluritest is accepted, because it is impossible to determine whether these cells would contribute to the germline. Germline transmission is still the gold standard for mouse iPS cells. We agree that the teratoma assay has issues.
4. Summary and outlook
We have discussed strategies for pluripotent stem cell quality control in general and the use of a novel global assay for pluripotency in human cells, termed PluriTest. This bioinformatic assay enables researcher to reliably test cell lines for pluripotency in an economical way, more rapidly that a teratoma assay, and without the sacrifice of lab animals. The results are much more informative than the conventional teratoma assay and as the associated microarray data is conveniently storable, the same samples can be reanalyzed with improved algorithms without any further lab or animal experimentation. In the near future, improvements and updates to PluriTest can be effortlessly distributed to all stem cell researchers worldwide through the online PluriTest platform.
With next generation sequencing technologies revolutionizing genome-wide analysis, we are developing new iterations of PluriTest to accommodate sequencing data. However, sequencing technologies have not yet matured to be as stable, reproducible, and inexpensive as microarray-based tools. Therefore, for the near future, microarrays will offer the best platform for obtaining reliable genome-wide information for diagnostic tests such as PluriTest.”
Acknowledgements
FJM is supported by an Else-Kröner Fresenius Stiftung fellowship. JFL is supported by CIRM (CL1-00502, RT1-01108, TR1-01250, RN2-00931-1), NIH (R33MH87925), the Millipore Foundation, and the Esther O’Keefe Foundation.
Reference
Last revised August 28, 2012. Published December 10, 2012. This chapter should be cited as: Müller, F.-J., Brändl, B., and Loring, J.F., Assessment of human pluripotent stem cells with PluriTest (December 10, 2012), StemBook, ed. The Stem Cell Research Community, StemBook, doi/10.3824/stembook.1.84.1, https://www.stembook.org.


